Fig. 2. Genome-wide CRISPR-Cas9 screen for regulators of NOXA-induced MCL1 degradation.
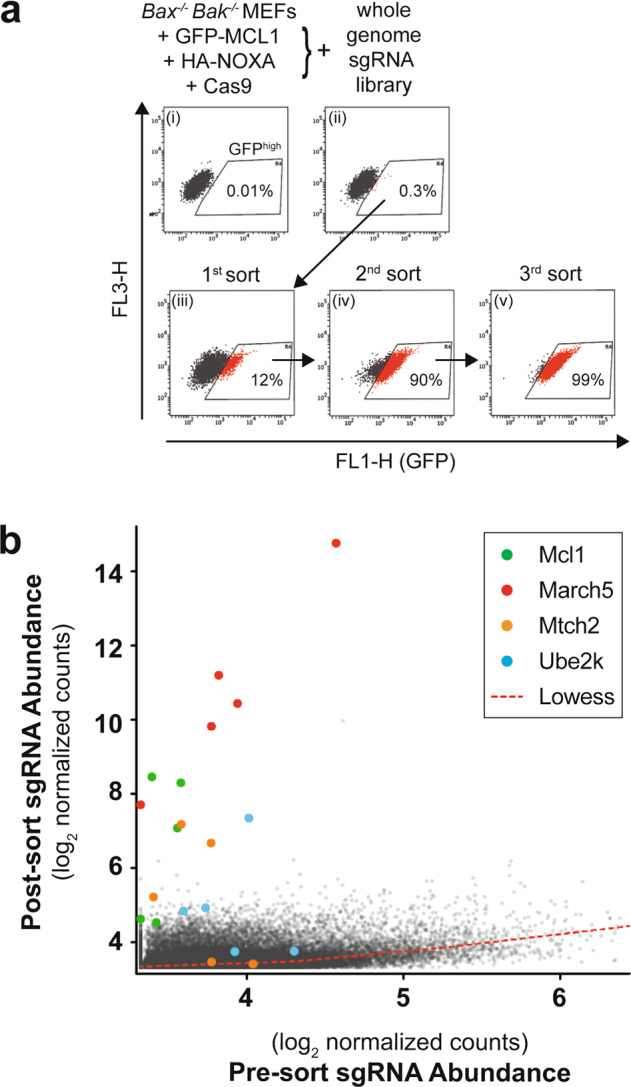
a Selection strategy to enrich cells in which the ability of NOXA to provoke MCL1 degradation has been disabled. Bax−/−Bak−/− MEFs engineered to express GFP-MCL1 along with HA-NOXA and Cas9 (i) were transduced with a genome-wide sgRNA lentivirus library [24]. Following transduction, a small proportion of GFPhigh cells was detected (ii) and enriched through three rounds of flow cytometric sorting (iii–v). b Quantitation of sgRNA abundance in the transduced cells before and after sorting. Multiple independent sgRNAs targeting Mcl1, March5, Mtch2, and Ube2k were enriched within the sorted GFPhigh cell population. These data are also summarized in Supplementary Table S3.
