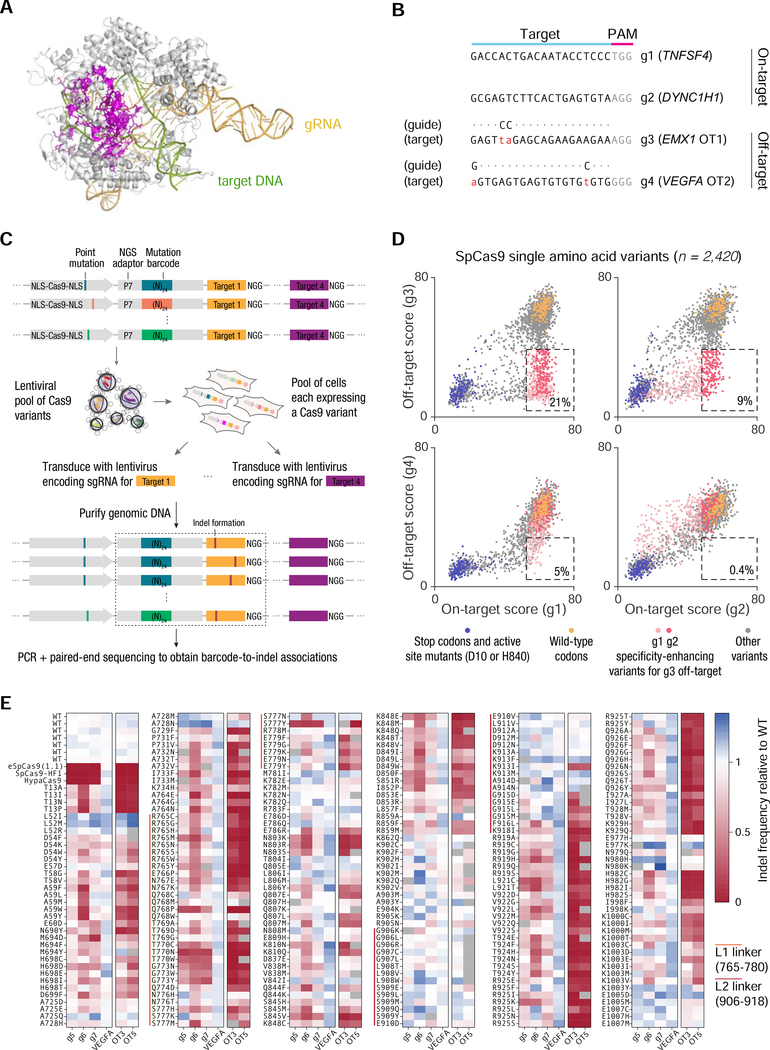
Figure 2 |. High-throughput profiling of SpCas9 mutant fitness in human cells.
(A) Crystal structure of SpCas9 (PDB ID: 5F9R) showing the positions of 157 residues (magenta) selected for mutagenesis. (B) Sequences of target sites used for screening. (C) Approach for pooled lentiviral screening of SpCas9 variants in HEK 293FT cells. (D) Scatter plots of on-target vs. off-target activity scores for 2,420 SpCas9 single amino acid variants. The dashed box in each subplot contains all variants with ≥80% of the median wild-type on-target activity and ≤50% of the median wild-type off-target activity; activities were calculated after subtracting the median background activity of stop codon variants. The percentage within each box represents the percentage of all variants that lie within the box. (E) On-target and off-target activity of 254 SpCas9 single amino acid variants, quantified by targeted deep sequencing of individually transfected constructs. See also Figure S2.