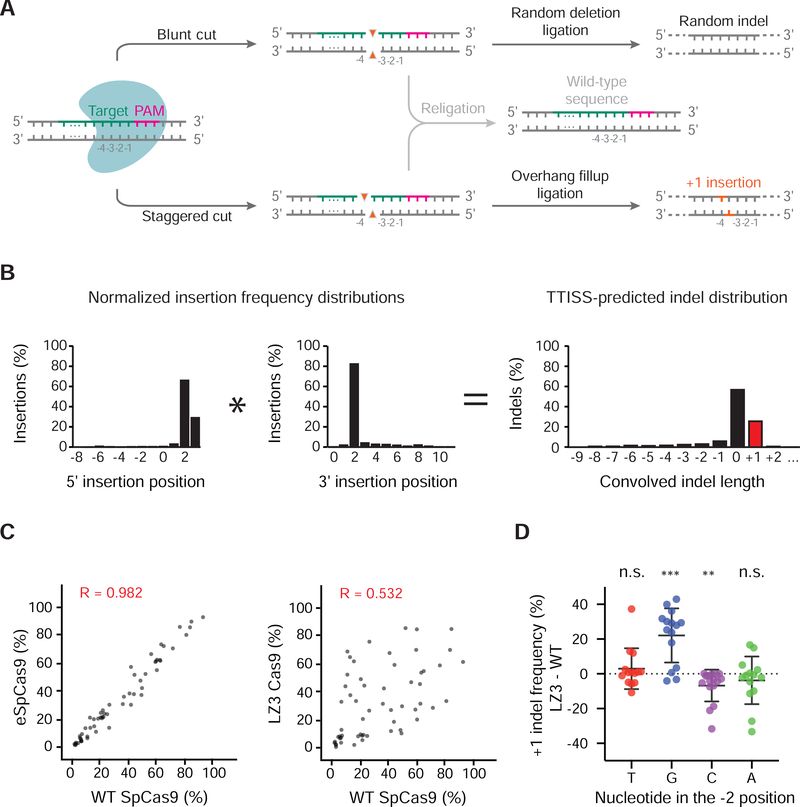
Figure 3 |. Multiplexed assessment of +1 indel frequencies using TTISS.
(a) Editing outcomes of nuclease-induced blunt or staggered cuts in the human genome. As a simplified model, blunt or staggered cuts can either be resected prior to re-ligation, creating random deletions (top panel) or re-ligated without resection (middle panel). Staggered 5’-overhangs can be filled in before re-ligation, causing duplication of base −4 respective to the PAM motif (bottom panel). (b) Schematic for convolution operation used to predict indel distributions by TTISS. (c) Representative examples of TTISS-predicted +1 insertion frequencies compared between specificity variants versus WT SpCas9 for 58 gRNAs. (d) Differential +1 indel frequencies between LZ3 Cas9 and WT SpCas9 +1 insertion frequencies from targeted indel sequencing, grouped by the nucleotide identity at the −2 position relative to the PAM. Results from two-tailed t-test for significant divergence from zero are indicated by ** (p < 0.01), *** (p < 0.001), n.s. (not significant). See also Figure S3.