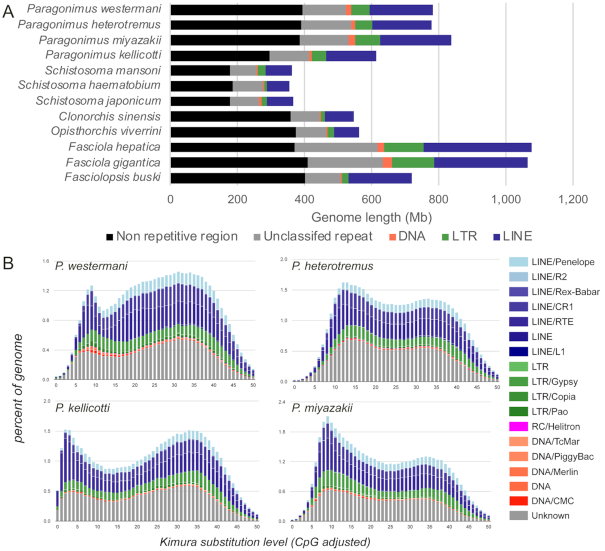
Figure 1:
Comparisons of the overall content of the assembled Paragonimus genome assemblies. Comparisons are based on (A) length (including statistics for other sequenced trematode genomes) and (B) repeat landscapes, measured using the Kimura substitution level, which indicates how much a repeat sequence has degenerated since its incorporation into the genome (i.e., how recently the repeat sequence was added). The high peak at the far left of P. kellicotti indicates a recent incorporation or active transposable element activity. LINE: long interspersed nuclear element; LTR: long terminal repeat.