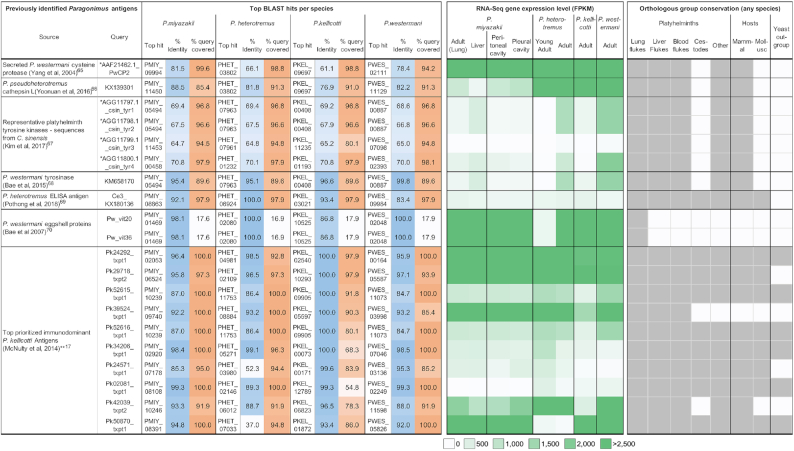
Figure 10:
Gene matches, expression level, and orthology for previously identified Paragonimus antigens. Top gene matches in each species (Diamond blastp) are shown, and the percent identity and percentage of the query sequence covered with the match are shown. Gene expression data correspond to the matched gene for each species, and orthology data indicate the conservation of the matched proteins according to the Orthologous Group analysis (dark grey = ortholog present in ≥1 species in group). *Query sequence was an amino acid sequence instead of a nucleotide sequence. **Of the top 25 P. kellicotti immunodominant antigen transcripts identified by McNulty and coworkers [17], the 10 best matches are presented (in terms of percent identity between the assembled transcript and the annotated gene). For the other 3 species, the BLAST searches were performed against the orthologous gene in P. kellicotti, not the original transcript sequence.