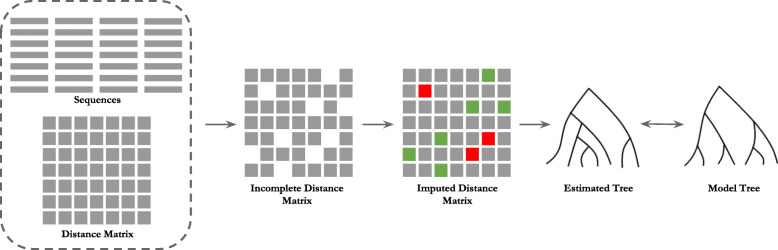
Fig. 1.
An overview of the experimental pipeline of this study. The input is either a set of sequences, or a complete distance matrix. We generate incomplete distance matrix from input sequences or input complete distance matrix by using various missingness mechanisms. Next, we apply various imputation techniques to impute the missing entries in the incomplete distance matrix and thereby, generating (complete) imputed distance matrices. Next, we estimate phylogenetic trees from the imputed distance matrices using FastME. Finally, we compare the estimated trees with the model tree to evaluate the performance of various imputation techniques