Abstract
Background & objectives:
Gall bladder cancer (GBC) is a fatal neoplasm, with a globally variable incidence rates. To improve the survival rate of patients, a newer set of biomarkers needs to be discovered for its early detection and better prognosis. Our earlier studies on GBC proteomics and whole-genome methylome data revealed expression of desmin to be significantly downregulated with correlated promoter hypermethylation during gall bladder carcinogenesis. Thus, to evaluate desmin as a potential biomarker for GBC, we carried out a detailed follow up study.
Methods:
Methylation-specific polymerase chain reaction (MS-PCR) (n=17, GBC and n=23, non-tumour control), real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) [n=14, GBC and n=14, adjacent non-tumour (ANT)], immunohistochemistry (n=27, GBC and n=14, non-tumour) and immunoblotting (n=13, GBC and n=13, ANT) were performed in surgically removed gall bladder tissue samples.
Results:
MS-PCR analysis showed methylation of desmin in 88.23 per cent (15/17) gall bladder tumour samples as compared to non-tumour tissues (39.13%, 9/23). Real-time qRT-PCR analysis revealed a significant downregulation of desmin expression in GBC as compared to ANT tissue. This was further confirmed by western blot, showing reduced expression of desmin protein in GBC, as compared to non-tumour tissue. Immunohistochemical analysis also showed a decreased level of desmin i.e., more than 95 per cent (26/27) in tumour cells compared to non-tumours (35.71%, 5/14).
Interpretation & conclusions:
The increased frequency of desmin promoter methylation which could be responsible for its significant downregulation, indicates its potential as a candidate biomarker for GBC. This requires further validation in a large group of patients to evaluate its clinical utility.
Keywords: Desmin, disease progression, DNA methylation, epigenetics, gall bladder cancer, molecular diagnostic
Gall bladder cancer (GBC) is the most common neoplasm of the biliary tract. Early detection of GBC is difficult due to the non-availability of well-defined set of specific biomarkers. At present, radical cholecystectomy is the only treatment strategy for resectable cancer that includes stage I, II and III cancers not spread outside the gall bladder. However, due to the lack of early detection tools, majority of gall bladder cancers are diagnosed at an advanced stage and unresectable cancers1. Better understanding of the molecular pathogenesis of GBC is important to identify biomarkers of diagnostic, therapeutic, and prognostic significance. Our earlier epidemiological studies revealed that gall bladder cancer-related morbidity was high in north central region of India2,3.
In our isobaric tags for relative and absolute quantitation (iTRAQ) based proteomic study of the gall bladder tumours and adjacent non-tumour (ANT) tissue4, we observed a 3.3-fold downregulation of desmin (DES) gene. Our genome-wide methylation analysis using Illumina's Infinium HumanMethylation450 BeadChip array5 revealed desmin to be hypermethylated in the promoter region in gall bladder tumours compared to non-tumours. Methylation-based dysregulation of genes is a hallmark of tumorigenesis and cancer progression6. Dysregulation of cytoskeletal proteins is one of the major events in tumorigenesis and tumour progression. Earlier studies proposed that desmin could be helpful in staging urothelial carcinomas and detecting invasion in muscularis propria7. The process of invasion and metastasis starts with the remodelling of cellular architecture, which also includes the interaction of cytoskeletal proteins. Intermediate filaments form a flexible but resilient framework that gives structural support to epithelium and functions in tissue repair and regeneration8. Desmin is a 52 kDa protein, composed of 470 amino acids and plays an essential structural role in the maintenance of muscle integrity. It is a main intermediate filament protein of type III in skeletal and cardiac muscle cells9,10. It resides at chromosome 2q35 and has a functional role in muscle contraction11. Desmin is reported to interact with several other cytoskeletal proteins (e.g., VIM, SYNC, SYNM, DSP, TCAP, TMOD1 and DMD), cancer-related proteins (e.g., NEB, S100B, S100A1 and SHBG) and many other functional proteins (e.g., CAPN1, AURKB, TRIM7, PKD1, ROCK1, SPTAN1 and AURKB)12,13. Immunostaining [immunohistochemistry (IHC)] for desmin showed a weak staining in more than 50 per cent of cells in leiomyosarcoma and positive staining in tubular epithelial cells of normal kidney14. In contrast, the gastrointestinal and extra-gastrointestinal stromal tumours cells showed a negative staining pattern for desmin15,16.
With collective evidence from the previous studies demonstrating the role of desmin in different cancer types and our previous global methylation analysis and proteomic study in gall bladder cancer, we hypothesized that this gene was downregulated during disease progression in gall bladder cancer through epigenetic mechanisms. To validate the hypothesis, we investigated the pattern of expression of desmin and its epigenetic regulation in gall bladder cancer patients. Our objective was to provide a proof of concept on the epigenetic silencing of desmin through promoter methylation in gall bladder cancer.
Material & Methods
The present study was conducted in the Centre for Genomics, Jiwaji University, Gwalior, India, after obtaining approval from the Institutional Human Ethics Committee. Written informed consent was taken from all the patients or their family members included in the present study.
Sample collection: Gall bladder cancer tissue and adjacent non-tumour tissue samples from patients undergoing radical cholecystectomy were collected at the department of Pathology, Cancer Hospital and Research Institute (CHRI), Gwalior, India, during 2007 to 2016. The patients were selected randomly based on their ultrasound findings about gall bladder lesions. Based on macroscopic findings post cholecystectomy, paired gall bladder tissue samples were excised during gross examination from tumour and ANT sites, and this was confirmed later with microscopic findings by the surgical pathologist (Table I). The excised tissue samples for molecular studies were frozen immediately at −80° C. Tissue microarray (TMA) was constructed using archival paraffin blocks of gall bladder cancer tissue and non-tumour tissue. For methylation-specific polymerase chain reaction (MS-PCR), 17 GBC, corresponding 17 ANT tissues and 6 cholecystitis tissues as non-tumour control, for real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR), 14 GBC and 14 ANT tissues, for immunoblotting 13 GBC and 13 ANT tissue samples and for IHC, 27 GBC and 14 non-tumour tissue samples were taken.
Table I.
Demographic details of the patients included in the study
| Characteristics | MS-PCR n (%) (n=23) |
Real-time PCR n (%) (n=14) |
IHC n (%) (n=27) |
Western blotting n (%) (n=13) |
|---|---|---|---|---|
| Gender | ||||
| Male | 5 (21.74) | 4 (28.57) | 6 (34.78) | 6 (46.16) |
| Female | 18 (78.26) | 10 (71.43) | 21 (65.22) | 7 (53.84) |
| Age (yr) | ||||
| ≤45 | 12 (52.17) | 2 (14.29) | 9 (33.33) | 4 (30.77) |
| >45 | 11 (47.83) | 12 (85.71) | 18 (66.67) | 9 (69.23) |
| Mean±SD (range) | 47.9±12.36 (22-80) | 52.71±9.13 (30-65) | 47.47±12.83 (26-72) | 51.92±10.69 (32-65) |
| Grades of cancer | ||||
| Grade I | 1 (5.88)* | 5 (35.71) | 3 (11.11) | 3 (23.08) |
| Grade II | 8 (47.06)* | 2 (14.29) | 9 (33.33) | 4 (30.77) |
| Grade III | 8 (47.06)* | 7 (50) | 15 (55.56) | 6 (46.15) |
| Location of ANT/non-tumour | ||||
| Neck | 19 (82.60) | 6 (42.86) | 13 (92.86)# | 6 (46.16) |
| Body | 2 (8.70) | 3 (21.43) | 0 (0)# | 2 (15.38) |
| Fundus | 2 (8.70) | 5 (35.71) | 1 (7.14)# | 5 (38.46) |
| Tumour (with/without stone)* | ||||
| With stone | 4 (23.53)* | 9 (64.29) | 6 (22.22) | 4 (30.77) |
| Without stone | 13 (76.47)* | 5 (35.71) | 21 (77.78) | 9 (69.23) |
*Information on pathological tumour grades, location of the tumour in gall bladder and the presence and/or absence of stones were available for 17 patients; #location available for 14 patients only. ANT, adjacent non-tumour; SD, standard deviation; IHC, immunohistochemistry; MS-PCR, methylation-specific polymerase chain reaction
Genomic DNA isolation and bisulphite modification: Genomic DNA (gDNA) was isolated from gall bladder tissue samples using QIAamp DNA mini kit (Qiagen, Germany) and quantitated by NanoDrop (Implen, Germany). The DNA quality was assessed on 0.8 per cent agarose gel. One microgram of DNA was taken for bisulphite modification using Zymo EZ DNA Methylation kit (Zymo Research, USA) following the manufacturer's protocol.
Detection of promoter methylation by methylation-specific (MS)-PCR: The following primer sequences were used for DES promoter for MS-PCR: for methylated sequence, forward primer: 5ˈ-TTTTTACGGTTACGGGTC-3ˈ; reverse primer: 5ˈ-CGTCGACACTATTTATATCCC-3ˈ and for unmethylated sequence, forward primer: 5ˈ-GT TTTTATGGTTATGGGTT-3ˈ; reverse primer: 5ˈ-CAT CAACACTATTTATATCCCT-3ˈ. The primers were designed using Methyl Primer Express® Software v1.0, (Thermo Fischer Scientific, USA). The PCR thermal conditions were as follows: initial denaturation at 95°C for 5 min, 35 cycles of denaturation 35 sec, annealing at 53°C for 30 sec, extension at 72°C for 45 sec and final extension for 10 min. The PCR products were resolved on 8% native polyacrylamide gel electrophoresis (PAGE). The MS-PCR was performed as described by Herman et al17 with some modifications.
RNA isolation and cDNA synthesis: Total RNA from gall bladder tissue samples (14 - tumour and 14 - non-tumour) was extracted using TRIzol solution (Qiagen, Hilden, Germany) and the RNeasy mini kit (Qiagen, Germany). Qiagen's cDNA synthesis kit was used to prepare cDNA following the manufacturer's protocol.
Real-time PCR: Real-time qRT-PCR11 (n=14, GBC and n=14, ANT) was performed using 0.75 μg of total RNA in technical duplicates for DES (Qiagen, Germany) and normalized to RRN18S (Qiagen, Germany) using QuantiTect SYBR Green RT-PCR kit (Qiagen, Germany) on the Applied Biosystems 7500, USA, platform. The data were graphically presented as a per cent fold change in tumour tissue normalized to that of ANT tissue.
Western blotting: Tissue samples corresponding to those used for real-time qRT-PCR were used for western blotting (n=14 GBC and n=14 ANT). Protein lysates were prepared from tissues in lysis buffer containing protease inhibitor cocktails (Roche Diagnostics, Germany) using a manual homogenizer and quantified using Bradford method18. Equal quantity of protein (30 μg) was loaded on a 110 per cent sodium dodecyl sulphate (SDS)-PAGE gel and transferred (blotted) onto a polyvinylidene fluoride (PVDF) membrane (GE Healthcare, USA). The membrane was blocked for 1 h at room temperature with blocking buffer [(5% non-fat skimmed milk in 1x Tris-buffered saline with 0.1% tween-20 (TBST)] and incubated overnight at 4°C with anti-desmin monoclonal antibody at a dilution of 1:1000 (Santa Cruz Biotechnology Inc., USA) and monoclonal anti-β-actin (Santa Cruz Biotechnology Inc., USA) at a dilution of 1:1000 in 1x TBST. Three washes of 1x TBST were given, followed by incubation with an anti-mouse, horseradish peroxidase (HRP)-conjugated secondary antibody dilution (1:10,000) at room temperature for two hours. The blot was then washed three times with 1x TBST and twice with 1x TBS. The blot was developed using a supersensitive enhanced chemiluminescence reagent (GE Healthcare, USA) according to the manufacturer's protocol on the BioRad Gel Doc System. Densitometry analysis was performed using the AlphaImager 2200 software (USA).
Tissue microarray and immunohistochemistry: TMA was constructed using paraffin sections (5 μm) of gall bladder tissue samples (GBC=27 and non-tumour tissues, n=14). The sections were deparaffinized in xylene and rehydrated in graded concentrations of alcohol (90, 70 and 30%). The slides were treated in a microwave at 80 watts in 10 mM citric acid (pH 6.0) for antigen retrieval, and endogenous peroxidase was quenched with 0.03 per cent H2O2 for 20 min. Non-specific binding was blocked by pre-incubation in blocking solution for one hour, and sections were incubated overnight at 4°C in a humidified chamber with a mouse monoclonal anti-desmin antibody (Santa Cruz Biotechnology Inc., USA), diluted to 1:500. The Vectastain Elite ABC kit (Vector Laboratories, USA) was used according to the manufacturers' instructions to visualize the signal. After incubation with primary antibody, sections were incubated with secondary antibody (Goat anti-mouse immunoglobulin G) conjugated with biotin-labelled polymer (Santa Cruz Biotechnology Inc., USA) for 60 min at room temperature, followed by incubation with avidin-biotin conjugates. The slides were washed in 1x phosphate-buffered saline, pH 7.4 (3 times for 5 min each) between each incubation step. Antibody detection was performed with 3,3'-diaminobenzidine (DAB) tetrahydrochloride peroxidase substrate, and tissues were counterstained with haematoxylin for one minute. The samples were dehydrated in 100 per cent ethanol and xylene and mounted with glass coverslip using DPX mountant for histology. The scoring (staining) was considered as follows: negative or no staining; 1+, weak positive, when less than 10 per cent of the cells were positive; 2+, positive, with 11-50 per cent positive cells, and 3+, strongly positive, with more than 50 per cent positive cells. The cellular localization of the immunostaining was also recorded.
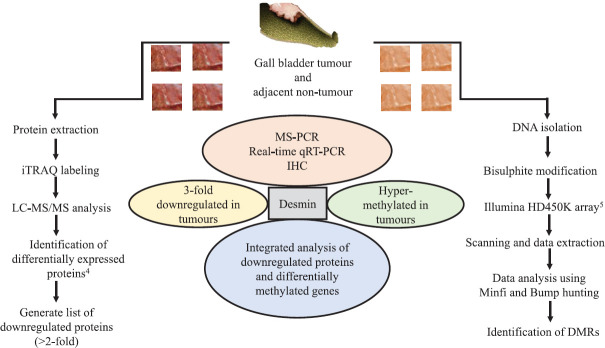
Figure 1 shows the investigations done in the present study.
Figure 1.
Workflow demonstrating systematic investigations leading to identification of desmin (DES) as an epigenetically regulated gene in gall bladder cancer. MS-PCR, methylation-specific polymerase chain reaction; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; IHC, immunohistochemistry; iTRAQ, isobaric tags for relative and absolute quantitation; LC-MS/MS, liquid chromatography-tandem mass spectrometry; DMRs, differentially methylated regions.
Statistical analysis: Student's t test and Fisher's exact test or Chi-square test were performed to test the significance of this study at 95 per cent confidence interval and 5 per cent level of significance using GraphPad Prism (www.graphpad.com). Pearson's Chi-square test was performed for categorical values. To analyze the difference between two data sets, two-tailed Wilcoxon rank-sum test or Student's t test was performed for real-time qRT-PCR and immunoblotting. Cohen's unweighted kappa and linearly weighted kappa tests were carried out to measure concordance of MS-PCR-based methylation and IHC-based expression of desmin.
Results
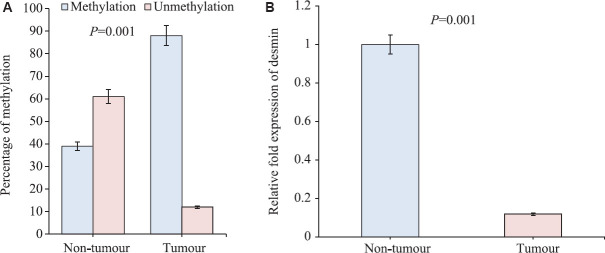
Methylation-specific PCR: Our previous whole-genome methylation analysis showed hypermethylation in 5' region (5'UTR) of desmin gene. The MS-PCR reconfirmed the methylation status of DES promoter, about 88 per cent (15/17) of GBC patients showed promoter hypermethylation (P=0.004) as compared to non-tumours, where only 39 per cent (9/23) showed promoter methylation (Fig. 2A). The studied cases comprised all grades of GBC (grade I, II and III) (Table I).
Fig. 2.
(A) Histogram showing statistical difference in methylation in tumours and non-tumours. (B) Relative fold change of desmin RNA expression by real-time polymerase chain reaction in gall bladder tumours and non-tumours. Fold change in tumour tissue normalized to non-tumour after subtracting the control 18S rRNA Ct values.
Real-time polymerase chain reaction (PCR): The real-time qRT-PCR results revealed that at transcript level, the expression of DES gene was significantly downregulated in gall bladder cancer tissues (P=0.001) as compared to its corresponding ANT tissues. All grades of GBC (grade I, II and III) in 12 of 12 GBC tissues showed downregulation of desmin (Fig. 2B and Table I).
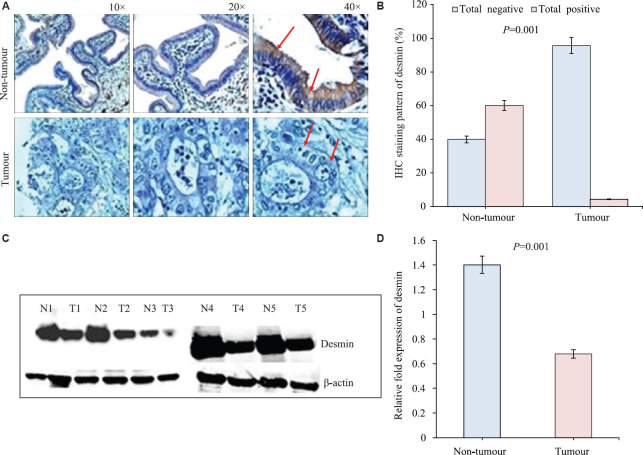
Protein expression [Western blot and immunohistochemistry (IHC)]: The western blot analysis revealed significant loss of expression of DES (P=0.0081) in GBC, compared to the corresponding ANT tissues (Fig. 3C and D). The result of western blot was reconfirmed by IHC of tumour and non-tumour tissues which showed adenocarcinoma cells to have significantly decreased level of desmin expression (26/27, 96.30%), compared to non-tumours (5/14, 35.71%) (P<0.001). It also revealed that the subcellular localization of desmin is cytoplasmic. Intense positive staining was observed in muscle cells of the gall bladder tissue (data not shown). The expression and staining patterns are shown in Figure 3A, B and Table II.
Fig. 3.
(A) Representative images of immunohistochemistry staining of desmin in gall bladder tumours (no diaminobenzidine, DAB stain) and adjacent non-tumour showing positive DAB stain, counterstain used was haematoxylin. (B) Histogram showing percentage of immunohistochemical staining patterns of desmin in gall bladder tumours and adjacent non-tumours. (C) Western blotting showing expression of desmin in five representative samples of gall bladder tumour (T1-T5) and their adjacent non-tumour (N1-N5). (D) Histogram showing the densitometric quantitation of desmin expression in gall bladder tumours and non-tumours from the western blotting (n=5 in both groups).
Table II.
Immunohistochemistry staining pattern (score) and subcellular localization of desmin protein in tumour and non-tumour cells
| Staining pattern | Non-tumour tissues | Tumour tissues |
|---|---|---|
| Total positive (1+, 2+ and 3+) | 9/14 | 1/27 |
| Total negative (0) | 5/14 | 26/27 |
| Subcellular localization | Cytoplasmic | |
| P | <0.001* | |
*Comparison between positive vs. negative in respective groups
Concordance of promoter methylation and protein expression: Cohen's kappa statistics weighs the frequencies of two methods and analyzes the proportions of agreement. The observed kappa of more than 80 per cent is considered as a strong agreement between the two methods19. The methylation of desmin by MS-PCR and downregulation of protein expression using IHC in tumours showed a 88.74 per cent (kappa value=0.7244) agreement [95% confidence interval (CI): 0.523-0.925] observed as a proportion of maximum possible agreement of methylation and downregulation of desmin in gall bladder cancer. The reverse was observed in non-tumour samples with no agreement of methylation and protein expression with 10 per cent (kappa value=0.0727) agreement (95% CI: 0-0.361).
Discussion
Our study revealed a significant downregulation of desmin in tumour tissue (GBC) as compared to ANT tissue associated with promoter methylation, suggesting epigenetic regulation of the gene during gall bladder tumorigenesis. The progression of cancer involves a complex cascade of molecular processes, and methylation-based gene silencing is a known mechanism among those. The cytoskeletal proteins are the major contributors towards the disorder strength, mainly due to their presence everywhere in the cell20. These structural proteins are the essential cellular components carrying out normal cellular functions. The disintegrity of these can lead to abnormal functioning of the cell. Desmin is one of the main components of the cytoskeletal system. It has been demonstrated that the phosphorylation of desmin in fasting and muscle atrophy interferes with its normal assembly process21. It has also been reported that the cytoplasmic presence of desmin protein in gall bladder tumour tissues may be due to the fact that mutant desmin causes collapsing of pre-existing desmin and aggregation in the cytoplasm9. Desmin has a major role in mitochondrial positioning and respiratory function22. Moreover, desmin has been identified as one of the hub genes for colon cancer by a network-based study23. Loss of expression of desmin has been reported in gastrointestinal stromal tumours24. The expression of desmin, a smooth muscle cell marker, is found to be reduced in dysplastic cells25. The intermediate filament may be considered as a useful diagnostic marker to identify neoplasm immunohistochemically, as these show co-expression of desmin, vimentin and keratin14. Intermediate filament has a suggestive role in signal transduction, which involves mechanosignaling in cells26. Our study finds further support from the report of Raparia et al27, who demonstrated a positive expression of desmin in gall bladder tissue samples, collected from chronic cholecystitis cases. Lower expression of desmin gene in myoblasts and myotube is also suggested to occur due to DNA methylation28. Like desmin, promoter methylation in some other genes, such as CDKN2A, ESR1, PGP9.5 and SSBP2, has also been observed in GBC29. DAPK1, DLC1, TIMP3 and RARβ2 were observed to be hypermethylated in tissues from cholecystitis to advance GBC stages30. It is presumed that desmin might be acting as a tumour suppressor gene, since hypermethylation in the promoter of any relevant gene may likely evolve novel tumour suppressor genes31.
Our earlier study on quantitative proteomics of GBC tissue identified desmin as a downregulated (3.3-fold) protein in gall bladder tumours compared to ANTs4. Poor expression of desmin is also observed in cholangiocarcinoma (cBioPortal, the Cancer Genome Atlas Data), while there is no mutation/deletion observed in the gene12,13. Our data showed significantly reduced expression of desmin in GBC tissues as compared to ANT tissues, mainly due to epigenetic alteration, and may not be due to deletion/mutation. In an earlier study on biliary tract cancer, a significant downregulation (−102-fold change in RNA expression) of desmin in gall bladder cancer was found11. The above studies, thus, suggest that epigenetics may have a significant role in the silencing of this gene, which is expressed poorly in the cancerous cells of the gall bladder. Our genome-wide methylome data5 revealed hypermethylation at 5ˈUTR region of desmin in GBC compared to ANT samples, which was corroborated by our current observation. The major limitation of this study was the sample size analyzed, which was because of infrequent availability of appropriately resected GBC tissue samples ideal for three different types of experiments qRT-PCT, western blotting and MS-PCR.
The present observations further reaffirmed our earlier proteomic and methylome studies, where desmin was found 3.3-fold downregulated at protein level and found to be methylated in gall bladder cancer. The available data from multiple studies32 provide convincing evidence to consider desmin as a potential biomarker for GBC that needs to be probed further with large sample size. Methylated desmin DNA may also be investigated as part of the circulatory tumour DNA in blood serum of a large group of calculus cholecystitis and various stages of gall bladder cancer to investigate the potential of methylated desmin DNA as a liquid biopsy tumour marker for gall bladder cancer. To further understand the functional implications and mechanisms involved in the dysregulation of desmin in gall bladder cancer, in vitro and in vivo experiments using cell lines and animal models should be employed.
Footnotes
Financial support & sponsorship: The last author (PKT) acknowledges the Madhya Pradesh Council of Science and Technology (MPCST) Bhopal, India, for providing funds to carry out this work. The first author (SB) was a recipient of fellowships provided by MPCST and Jiwaji University, Gwalior, India. Authors also acknowledge the University Grants Commission-Special Assistance Programme (UGC-SAP) and the Department of Science and Technology (DST), Government of India, for providing partial financial support in conducting this work
Conflicts of Interest: None.
References
- 1.Andrén-Sandberg A. Diagnosis and management of gallbladder cancer. N Am J Med Sci. 2012;4:293–9. doi: 10.4103/1947-2714.98586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbhuiya MA, Singh TD, Gupta S, Shrivastav BR, Tiwari PK. Incidence of gall bladder cancer in rural and semi-urban population of North Central India: A first insight. Internet J Epidemiol. 2009;7:1820–2155. [Google Scholar]
- 3.Barbhuiya MA, Singh TD, Poojary SS, Gupta S, Kakkar M, Shrivastav BR, et al. Gallbladder cancer incidence in Gwalior district of India: Five-year trend based on the registry of a regional cancer center. Indian J Cancer. 2015;52:430–7. doi: 10.4103/0019-509X.176736. [DOI] [PubMed] [Google Scholar]
- 4.Sahasrabuddhe NA, Barbhuiya MA, Bhunia S, Subbannayya T, Gowda H, Advani J, et al. Identification of prosaposin and transgelin as potential biomarkers for gallbladder cancer using quantitative proteomics. Biochem Biophys Res Commun. 2014;446:863–9. doi: 10.1016/j.bbrc.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma P, Bhunia S, Poojary SS, Tekcham DS, Barbhuiya MA, Gupta S, et al. Global methylation profiling to identify epigenetic signature of gallbladder cancer and gallstone disease. Tumour Biol. 2016;37:14687–99. doi: 10.1007/s13277-016-5355-9. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe Y, Maekawa M. Methylation of DNA in cancer. Adv Clin Chem. 2010;52:145–67. doi: 10.1016/s0065-2423(10)52006-7. [DOI] [PubMed] [Google Scholar]
- 7.Saha K, Saha A, Datta C, Chatterjee U, Ray S, Bera M. Does desmin immunohistochemistry have a role in assessing stage of urothelial carcinoma in transurethral resection of bladder tumour specimens? Clin Cancer Investig J. 2014;3:502–7. [Google Scholar]
- 8.Paulin D, Li Z. Desmin: A major intermediate filament protein essential for the structural integrity and function of muscle. Exp Cell Res. 2004;301:1–7. doi: 10.1016/j.yexcr.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Schröder R, Goudeau B, Simon MC, Fischer D, Eggermann T, Clemen CS, et al. On noxious desmin: Functional effects of a novel heterozygous desmin insertion mutation on the extrasarcomeric desmin cytoskeleton and mitochondria. Hum Mol Genet. 2003;12:657–69. doi: 10.1093/hmg/ddg060. [DOI] [PubMed] [Google Scholar]
- 10.Mericskay M, Parlakian A, Porteu A, Dandré F, Bonnet J, Paulin D, et al. An overlapping CArG/octamer element is required for regulation of desmin gene transcription in arterial smooth muscle cells. Dev Biol. 2000;226:192–208. doi: 10.1006/dbio.2000.9865. [DOI] [PubMed] [Google Scholar]
- 11.Miller G, Socci ND, Dhall D, D'Angelica M, DeMatteo RP, Allen PJ, et al. Genome wide analysis and clinical correlation of chromosomal and transcriptional mutations in cancers of the biliary tract. J Exp Clin Cancer Res. 2009;28:62. doi: 10.1186/1756-9966-28-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the BioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desnoyers MM, Haines DM, Searcy GP. Immunohistochemical detection of intermediate filament proteins in formalin fixed normal and neoplastic canine tissues. Can J Vet Res. 1990;54:360–5. [PMC free article] [PubMed] [Google Scholar]
- 15.Li FP, Fletcher JA, Heinrich MC, Garber JE, Sallan SE, Curiel-Lewandrowski C, et al. Familial gastrointestinal stromal tumor syndrome: Phenotypic and molecular features in a kindred. J Clin Oncol. 2005;23:2735–43. doi: 10.1200/JCO.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Li ZY, Huan XQ, Liang XJ, Li ZS, Tan AZ. Clinicopathological and immunohistochemical study of extra-gastrointestinal stromal tumors arising from the omentum and mesentery. Zhonghua Bing Li Xue Za Zhi. 2005;34:11–4. [PubMed] [Google Scholar]
- 17.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 19.McHugh ML. Interrater reliability: The kappa statistic. Biochem Med (Zagreb) 2012;22:276–82. [PMC free article] [PubMed] [Google Scholar]
- 20.Damania D, Subramanian H, Tiwari AK, Stypula Y, Kunte D, Pradhan P, et al. Role of cytoskeleton in controlling the disorder strength of cellular nanoscale architecture. Biophys J. 2010;99:989–96. doi: 10.1016/j.bpj.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker LK, Gillis DC, Sharma S, Ambrus A, Herrmann H, Conover GM. Nebulin binding impedes mutant desmin filament assembly. Mol Biol Cell. 2013;24:1918–32. doi: 10.1091/mbc.E12-11-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milner DJ, Mavroidis M, Weisleder N, Capetanaki Y. Desmin cytoskeleton linked to muscle mitochondrial distribution and respiratory function. J Cell Biol. 2000;150:1283–98. doi: 10.1083/jcb.150.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang W, Li X, Rao S, Wang L, Du L, Li C, et al. Constructing disease-specific gene networks using pair-wise relevance metric: Application to colon cancer identifies interleukin 8, desmin and enolase 1 as the central elements. BMC Syst Biol. 2008;2:72. doi: 10.1186/1752-0509-2-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JK, Choi SH, Lee S, Min KO, Yun SS, Jeon HM. Malignant gastrointestinal stromal tumor of the gallbladder. J Korean Med Sci. 2004;19:763–7. doi: 10.3346/jkms.2004.19.5.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong YC, Tam NNC. Dedifferentiation of stromal smooth muscle as a factor in prostate carcinogenesis. Differentiation. 2002;70:633–45. doi: 10.1046/j.1432-0436.2002.700916.x. [DOI] [PubMed] [Google Scholar]
- 26.Lowery J, Kuczmarski ER, Herrmann H, Goldman RD. Intermediate filaments play a pivotal role in regulating cell architecture and function. J Biol Chem. 2015;290:17145–53. doi: 10.1074/jbc.R115.640359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raparia K, Zhai QJ, Schwartz MR, Shen SS, Ayala AG, Ro JY. Muscularis mucosae versus muscularis propria in gallbladder, cystic duct, and common bile duct: Smoothelin and desmin immunohistochemical study. Ann Diagn Pathol. 2010;14:408–12. doi: 10.1016/j.anndiagpath.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 28.Lindahl Allen M, Koch CM, Clelland GK, Dunham I, Antoniou M. DNA methylation-histone modification relationships across the desmin locus in human primary cells. BMC Mol Biol. 2009;10:51. doi: 10.1186/1471-2199-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kagohara LT, Schussel JL, Subbannayya T, Sahasrabuddhe N, Lebron C, Brait M, et al. Global and gene-specific DNA methylation pattern discriminates cholecystitis from gallbladder cancer patients in Chile. Future Oncol. 2015;11:233–49. doi: 10.2217/fon.14.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.García P, Manterola C, Araya JC, Villaseca M, Guzmán P, Sanhueza A, et al. Promoter methylation profile in preneoplastic and neoplastic gallbladder lesions. Mol Carcinog. 2009;48:79–89. doi: 10.1002/mc.20457. [DOI] [PubMed] [Google Scholar]
- 31.Goldin RD, Roa JC. Gallbladder cancer: A morphological and molecular update. Histopathology. 2009;55:218–29. doi: 10.1111/j.1365-2559.2008.03192.x. [DOI] [PubMed] [Google Scholar]
- 32.Altmannsberger M, Weber K, Droste R, Osborn M. Desmin is a specific marker for rhabdomyosarcomas of human and rat origin. Am J Pathol. 1985;118:85–95. [PMC free article] [PubMed] [Google Scholar]