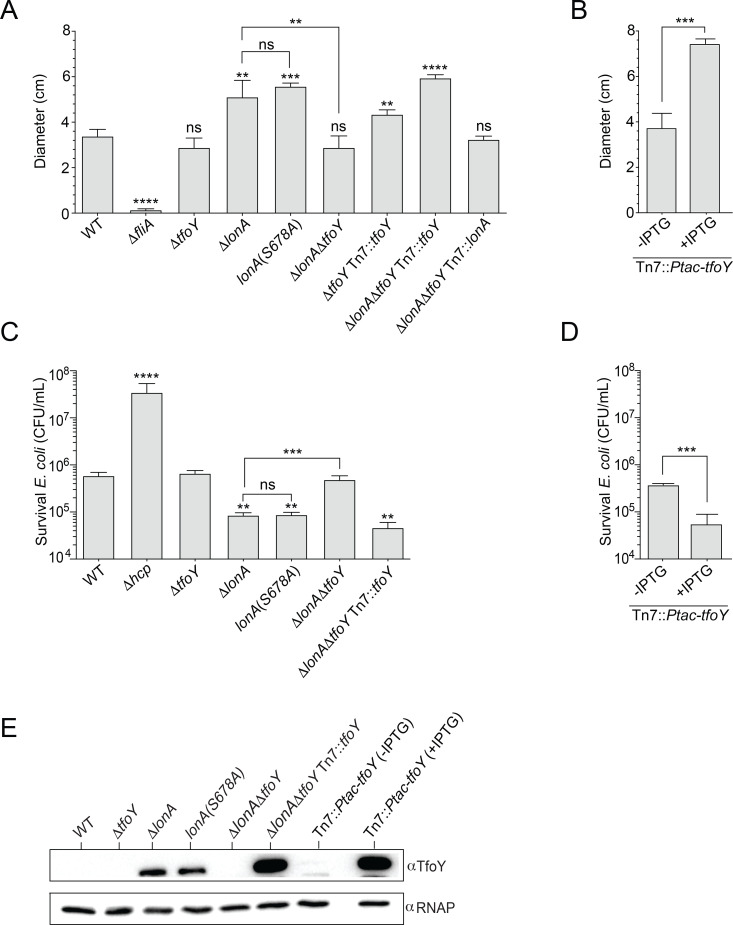
Fig 3. LonA represses motility and the T6SS through TfoY.
Quantification of flagellar motility and T6SS killing experiments. For motility assays, single colonies were stabbed into LB soft agar plates (0.3% agar) and incubated at 30°C for approximately 18 hours. (A) Swimming motility phenotypes of WT, ΔfliA (negative control) and various ΔlonA and/or ΔtfoY deletions as well as their complementation strains from the Tn7 site. (B) Overexpression of tfoY from the Ptac promoter in plates with and without IPTG. (C) The T6SS killing phenotypes of various tfoY and lonA deletion mutants as well as their complementation strains were analyzed. T6SS killing was determined by enumerating the survival of E. coli strain MC4100, which is susceptible to T6SS attack. In addition, hcp was included as a negative control for T6SS dependent killing and lonA(S678A) as a control for LonA-dependent proteolysis. (D) Overexpression of tfoY from the Ptac promoter on plates with or without IPTG. Motility and T6SS-dependent killing experiments represent the average and SD of at least three independent experiments. Statistical analysis was performed using an unpaired Student’s t-test. Statistical values indicated are (**p<0.01, ***p < .001, and ****p < .0001). (E) Abundance of natively produced TfoY as well as overexpressed TfoY from the Ptac promoter.