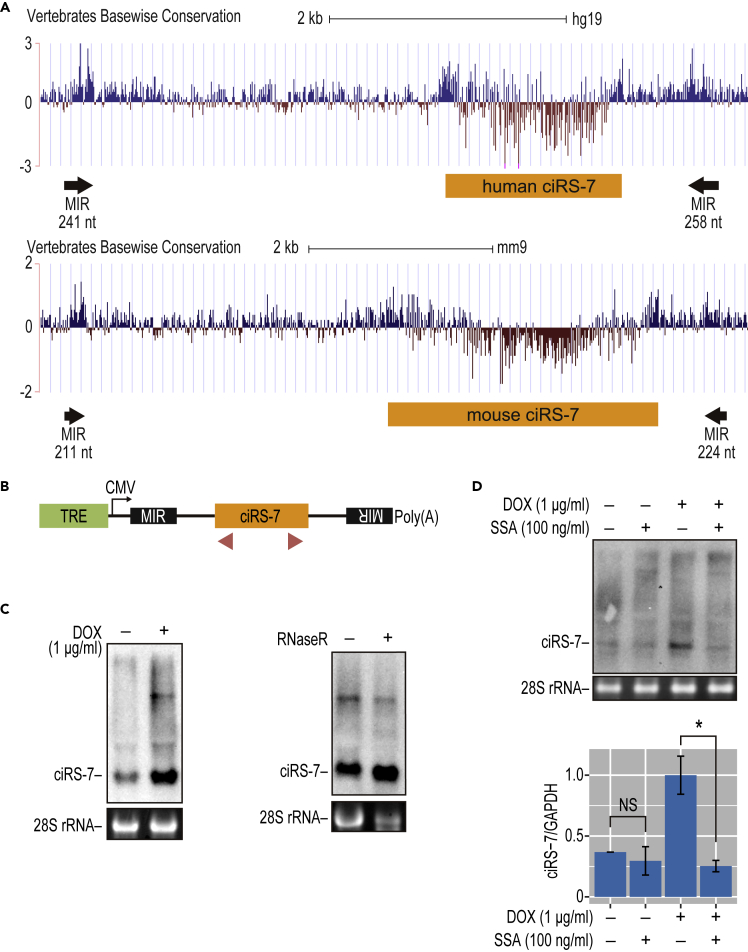
Figure 1.
The Production of ciRS-7 Depends on Inverted MIRs in the Flanking Regions
(A) The ciRS-7 locus (human/mouse ciRS-7) and the identified conserved inverted MIR elements (MIRs with the lengths are indicated). The complementarity of these inverted MIRs was modeled in Figure S8. The vertebrate base-wise conservations of human (upper) and mouse (lower) genomic sequences around the ciRS-7 precursor are displayed in the UCSC Genome Browser (GRCh37/hg19 version).
(B) Schematic structure of ciRS-7-expression plasmids. Transcription is driven by a CMV promoter, and an arrow denotes the initiation site. The tetracycline response element (TRE), inverted MIR elements, ciRS-7 coding exon (ciRS-7) are indicated. Red triangle indicates the positions of PCR primers used to detect ciRS-7 and its precursor, respectively.
(C) Detection of ciRS-7 from expression plasmid in stably transduced HEK293 cells. The cells were treated with DOX and extracted total RNA was analyzed by Northern blotting (left panel). 28S rRNA was visualized by ethidium bromide staining as a loading control. To validate the circular structure of ciRS-7, total RNA was treated with RNase R prior to the Northern blotting analysis (right panel).
(D) The effect of splicing inhibitor, SSA, on the production of ciRS-7. The same stably transduced cells described in (C) were treated with DOX (+lanes) followed by SSA addition (+lanes). After 12 h culture, extracted total RNA was analyzed by Northern blotting to detect ciRS-7 (upper panel). Ethidium bromide-stained 28S rRNA was shown as a loading control. The total RNA was also analyzed by RT-qPCR, and the graph shows quantification of the ciRS-7 generation (lower panel). ciRS-7 expression was normalized to GAPDH (ciRS-7/GAPDH) and plotted as ratios to the value of DOX-induced cells. Means ± SD are given for three independent experiments (∗p < 0.05, NS = not significant).