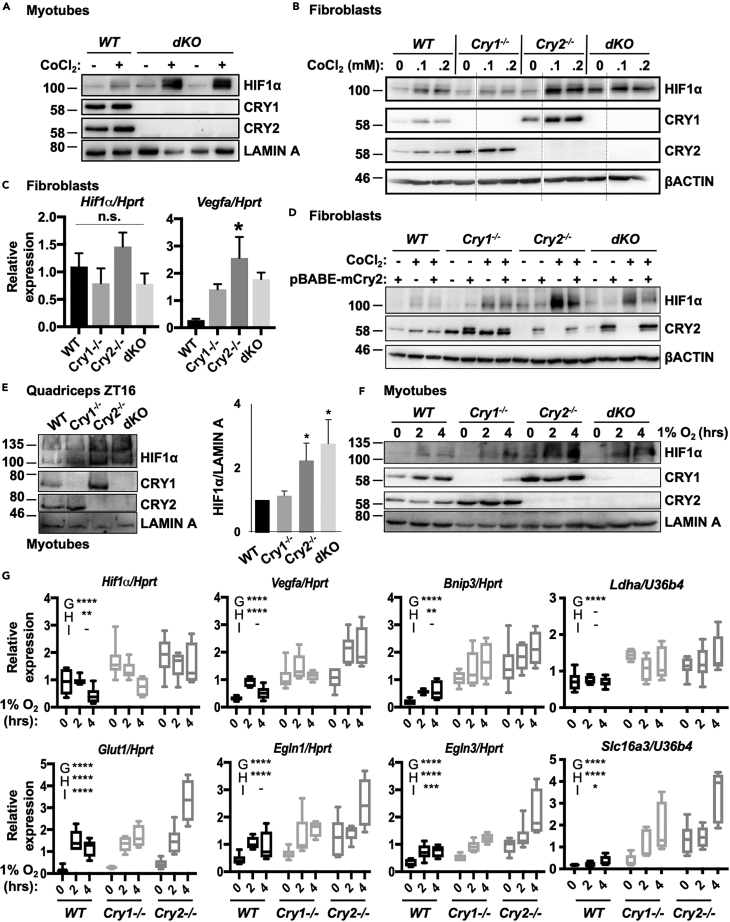
Figure 1.
CRYs Suppress HIF1α
(A) Accumulation of HIF1α protein detected by immunoblot (IB) in unsynchronized primary myotubes (1°MTs) isolated from WT and dKO mice and treated with vehicle control or 100 μM CoCl2. Two technical replicates are shown for dKO 1°MTs.
(B) Accumulation of HIF1α protein detected by IB in ear fibroblasts (EFs) isolated from WT, Cry1−/−, Cry2−/−, and dKO mice and treated with vehicle control or 0–0.2 mM CoCl2. Faint vertical lines indicate where blot images were spliced to remove samples treated with 50 μM CoCl2.
(C) Expression of the indicated transcripts measured by quantitative PCR (qPCR) in fibroblasts of the indicated genotypes, normalized to Hprt.
(D) Accumulation of HIF1α protein detected by IB in EFs isolated from mice of the indicated genotype and infected with virus carrying either empty vector or pBABE-mCry2 plasmid, before treatment with vehicle control (−) or 100 μM CoCl2 (+). (Note: the faint CRY2 signal in lane 12 reflects a small amount of sample from lane 13 that spilled into the neighboring well).
(E) Left, accumulation of HIF1α protein detected by IB in nuclei of cells isolated from quadriceps muscles of mice of the indicated genotype at ZT16. Right, quantitation of three experiments performed as shown at left.
(F) Accumulation of HIF1α protein detected by IB in unsynchronized 1°MTs isolated from mice of the indicated genotype upon exposure to 1% O2 for 0–4 h.
(G) Expression of the indicated transcripts measured by qPCR in unsynchronized 1°MTs plated under parallel conditions as those in (F). For (C and E), data represent the mean + SD for 3 samples per condition. ∗p < 0.05 versus WT by one-way ANOVA with Dunn's multiple comparison test. In (G) data represent the range (min to max with mean ± SD shown in box) for 6 samples per condition, each measured in triplicate. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 for a main effect of genotype (G), hypoxia (H), or an interaction between the two, determined by two-way ANOVA. Results of post-hoc analysis are not shown to highlight the main effect results of two-way ANOVA. The position of molecular weight markers (in kDa) are shown to the left of all western blot images. Note that overexpressed epitope-tagged proteins are slightly larger than endogenous ones.
See also Figure S1.