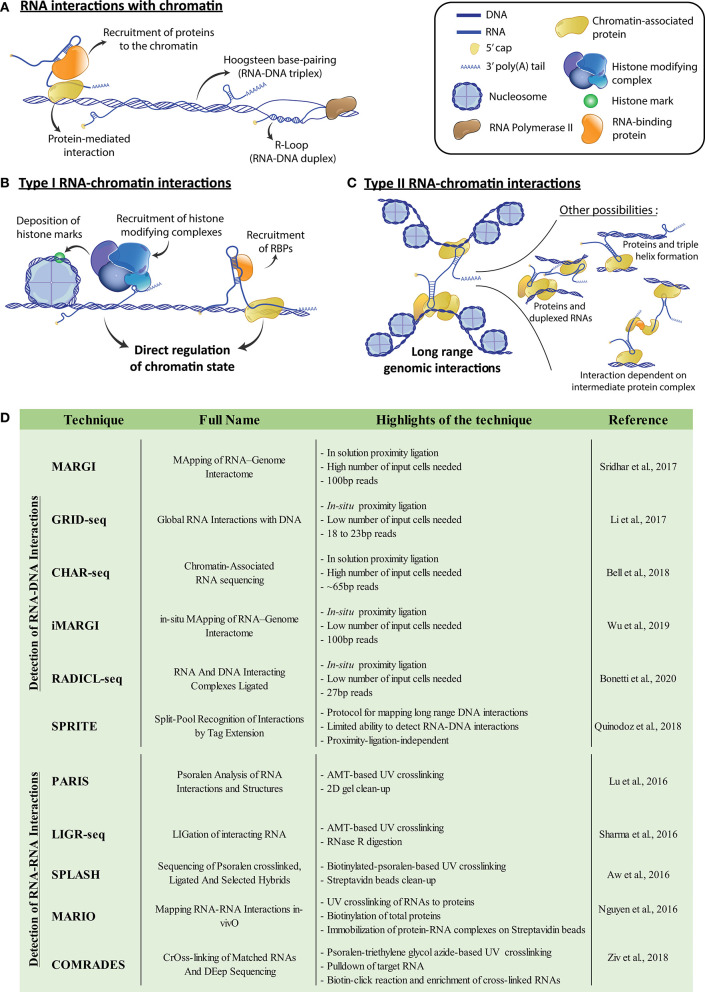
Figure 1.
(A) Potential means of chromatin-RNA association. Interactions can be mediated by protein intermediates or rely on either Hoogsteen base-pairing to form RNA-DNA triplexes, or Watson-Crick base-pairing to form RNA-DNA duplexes such as R-loops. (B) Representation of potential type I RNA-chromatin interactions, where RNAs recruit associated proteins to the chromatin, enabling the modulation of genomic accessibility. For example, recruited proteins can be part of chromatin modifying complexes or act as transcription factors. (C) Representation of potential type II RNA-chromatin interactions, where RNAs interact with genetically distant loci and modulate genome architecture, potentially bringing them physically closer. Interactions can be direct associations of the RNA with the DNA, or be mediated either by chromatin-bound proteins, or RNA duplexing. RNA bound protein complex intermediates can also generate indirect interactions. (D) Table of referenced techniques for identification of genome and transcriptome-wide DNA-RNA or RNA-RNA interactions, respectively. For more details, we refer the reader to the corresponding publications within the table. AMT, 4′-aminomethyl-4,5′,8-trimethylpsoralen.