Figure 2. Biological Characterization of protein and non-protein transcripts.
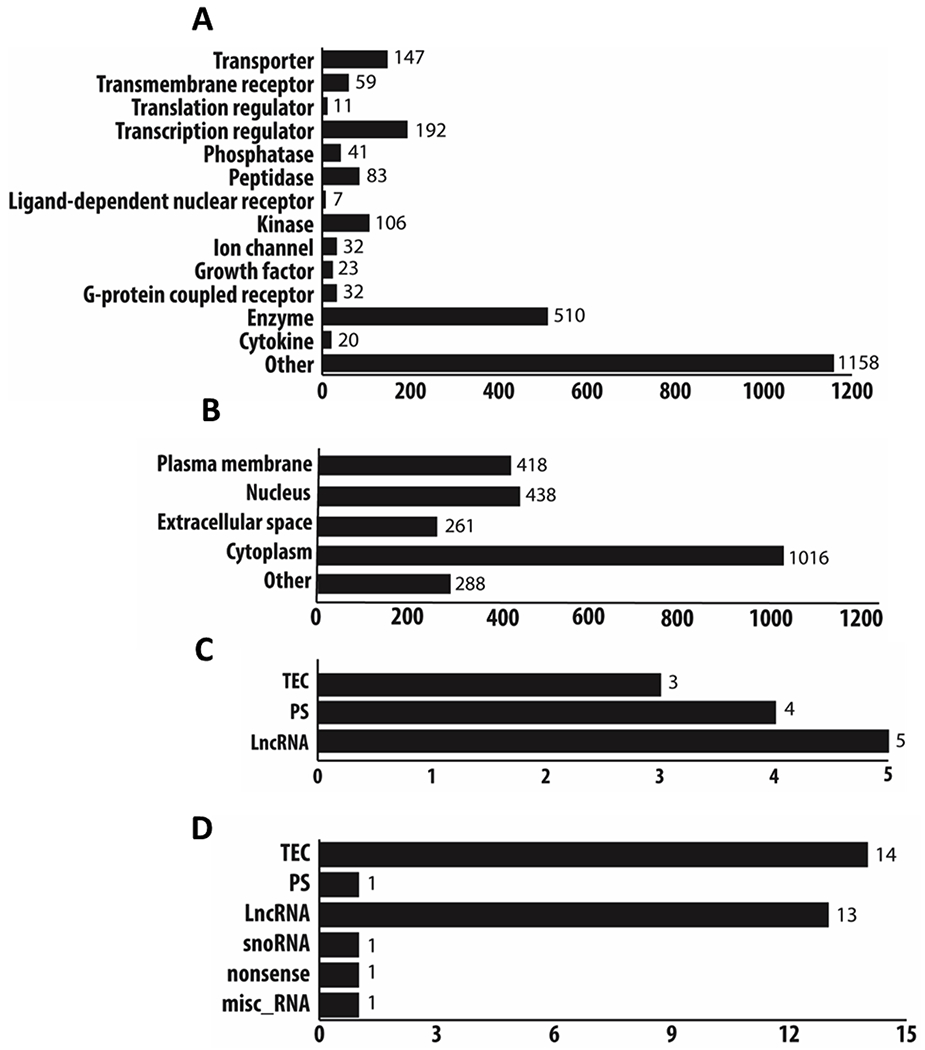
All differentially expressed protein coding transcripts (CT1 as compared to CT23) with a p-value < 0.05 and a fold change ≥ 2.0 (edgeR, v3.24.1, B-H adjustment) were categorized into different class types and location of action in the cell via the ingenuity pathway analysis database (v48207413; a and b). A large majority of protein coding transcripts were characterized into enzymes, kinases, transcriptional regulators, and transporters (a). Cellular localization of the protein-coding transcripts was identified to be expressed in the cytoplasm, nucleus, and plasma membrane (b). Using the datamining tool Biomart (Ensembl release 98), we also classified the non-protein coding differentially expressed transcripts (p < 0.05, fold change ≥±2.0, edgeR, v3.24.1, B-H adjustment) into different categories (c and d). Non-protein coding transcripts that were positively differentially expressed had transcripts that were classified as: TEC, PS, and LncRNA (c). Non-protein coding transcripts that were negatively differentially expressed were classified as: TEC, PS, LncRNA, snoRNA, nonsense, and misc RNA (d). TEC=To be experimentally confirmed; PS=pseudogene; LncRNA=long non-coding RNA. Note: definitions of specific biotypes for each transcript can be found on the Ensembl website for further clarification (https://useast.ensembl.org/info/genome/genebuild/biotypes.html).
