Figure 3. Gene ontology analysis of the protein coding mouse transcriptome.
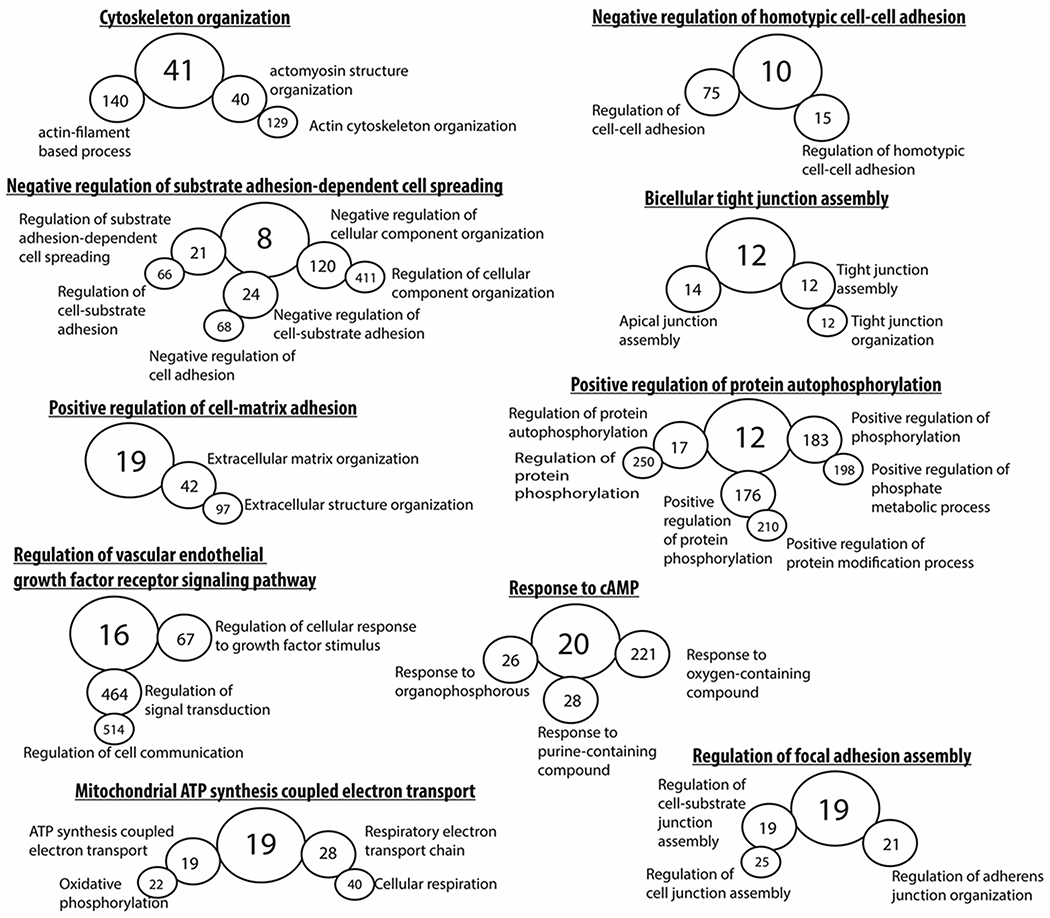
Gene ontology hierarchal analysis of all differentially expressed (CT1 as compared to CT23) protein coding transcripts (p < 0.05, fold change ≥±2.0, edgeR, v3.24.1, B-H adjustment) revealed terms that were consistent to previously known processes involved in RPE phagocytosis such as cell-cell adhesion, tight junction formation, focal adhesion assembly, protein autophosphorylation, and cAMP signaling. Interestingly, the identification of mitochondrial respiration has not been previously demonstrated before in the context of RPE phagocytosis in vivo. Greater circle diameter indicative of parent term in gene ontology hierarchal analysis. The number in each circle represents the exact number of genes that make up that gene ontology term identified in the current study’s dataset.
