Figure 4. RPE associated phagocytic pathways under circadian control.
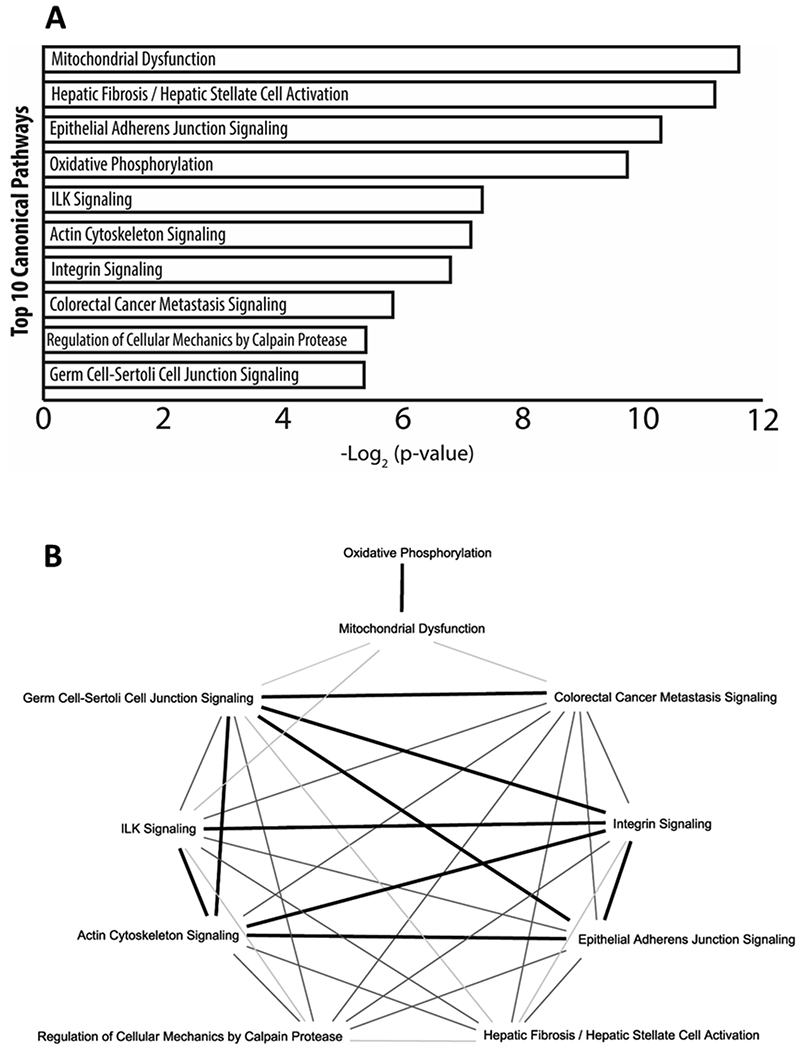
Subjecting all differentially expressed (CT1 as compared to CT23) protein-coding transcripts (edgeR, v3.24.1, B-H adjustment; p < 0.05, fold change ≥ ±2.0) to canonical pathway analysis with IPA (Ingenuity pathway analysis v48207413) revealed previously known biological pathways associated with RPE phagocytosis such as: integrin signaling, actin cytoskeleton signaling, germ cell-sertoli cell junction signaling, and epithelial adherens junction signaling. Much like the gene ontology analysis (Figure 3), the identification of mitochondrial processes was also apparent in the canonical pathway analysis. When the top 10 canonical pathways were plotted for common genes shared among the pathways, it revealed that integrin signaling, ILK signaling, actin cytoskeleton signaling, germ cell-sertoli cell junction signaling, and epithelial adherens junction signaling all had at least 20 genes in common (dark bold line) among them suggesting strong cross talk in these different pathways in mediating the process of RPE phagocytosis (b). While biological processes such as regulation of cellular mechanics by calpain protease and colorectal cancer metastasis signaling had less than 20 genes in common to the other identified pathways (light and medium grey lines, b). Interestingly, while mitochondrial processes were highly expressed in both gene ontology (Figure 3) and in canonical pathway analysis, there is very little overlap (light grey lines) among the other phagocytic associated pathways identified (b).
