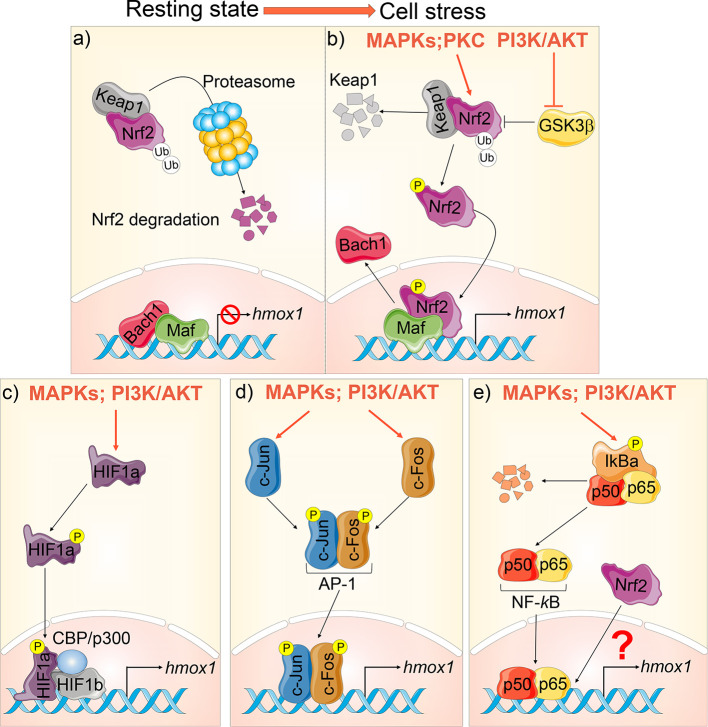
Figure 2.
Hmox1 transcriptional regulation. (A) At steady state, Nrf2 is bound to Keap1 in the cytoplasm and targeted for proteasomal degradation. Bach‐1 is bound to Maf at the promoter region of the hmox1, suppressing its transcription. (B) During cellular stress, hmox1 expression is activated in several ways: mitogen-activated protein kinase (MAPKs) and protein kinase C (PKC) phosphorylate Nrf2. This stabilizes Nrf2, leading to its translocation into the nucleus. PI3K/AKT inhibits GSK3β. When activated, GSK3β facilitates the ubiquitination and proteasomal degradation of Nrf2. Once in the nucleus, Nrf2 displaces Bach‐1 at the hmox1 promoter leading to transcription. (C) HIF-1 is a heterodimer composed of HIF-1α and HIF-1β. HIF-1α phosphorylation leads to its translocation to the nucleus and association to HIF-1β and CBP/p300, thus inducing hmox1 transcription. (D) Phosphorylation of c-Fos and c-Jun leads to their translocation to the nucleus and formation of the AP-1 complex, which induces hmox1 expression; (E) NF-κB is sequestered in the cytosol under basal conditions by the inhibitor IκB. Phosphorylation results in the proteasomal degradation of IĸB and the consequent release and nuclear translocation of NF-κB dimers (p50/p65) which targets hmox1 activation. A complex crosstalk between NF-κB and Nrf2 can also inhibit hmox1 transcription.