Figure 1.
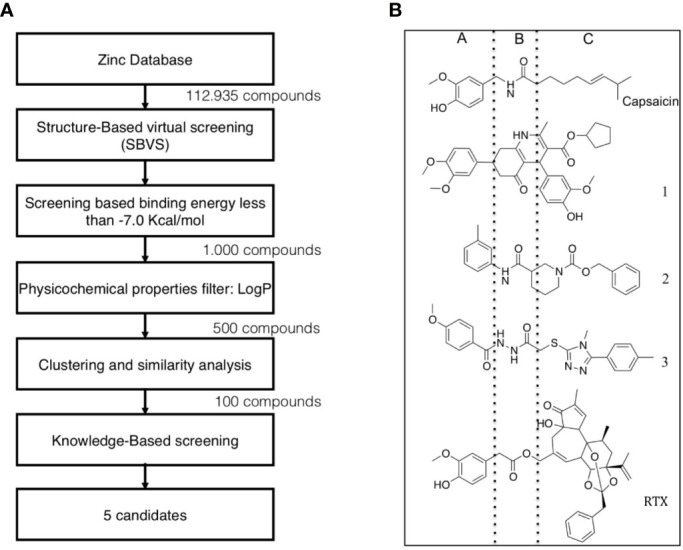
Workflow of TRPV1 Virtual Screening. (A) A SBVS over a ZINC Database with 112,935 compounds was performed using the TRPV1 channel structure as target. Results were filtered by excluding all molecules with binding energy below −7 kcal/mol, which reduced the candidate molecules to 1,000. A restriction by LogP values compatible with the ADME criteria was subsequently applied, followed by clustering and bibliography-based selection, which rendered five potential candidate molecules. (B) Structural comparison between capsaicin, the novel TRPV1 channel agonists found in this study and Resiniferatoxin. The molecules were decomposed into three regions, called A, B, and C. A-region comprises the Vanilloid ring, B-Region comprises a linker containing the electron donor–acceptor pair and C-region represents the hydrophobic region of each molecule.
