Table 1.
Theoretical parameters obtained in silico for each compound.
| Compound | Number | Structure | EA Kcal/mol | LogP | EC50 (nM) |
|---|---|---|---|---|---|
| Capsaicin |
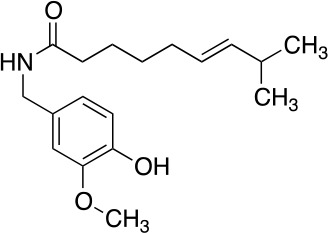
|
-7.0 | 3.58 | 440 ± 66 | |
| Compound | 1 |
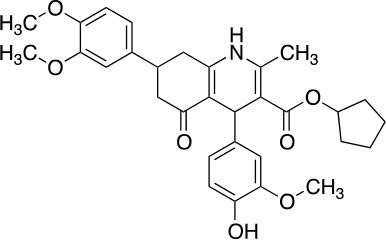
|
-9.3 | 4.65 | 53 ± 6 |
| Compound | 2 |
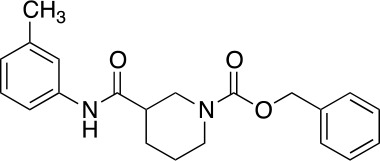
|
-9.2 | 3.31 | 53 ± 4.3 |
| Compound | 3 |
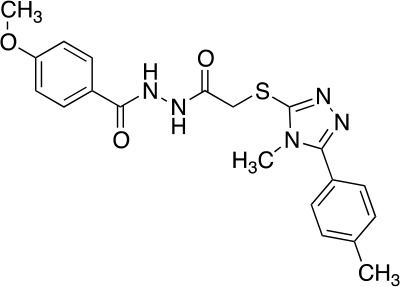
|
-9.1 | 2.78 | 92 ± 10 |
| Resiniferatoxin |
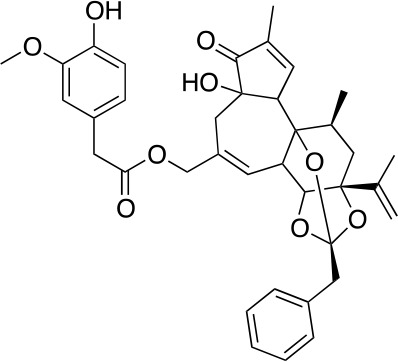
|
-9.0 | 4.51 | 0.7 ± 3 |
Structure (column 3), potential binding energy (column 4), theoretical Octanol/water partition coefficient (LogP; column 5) and empirical EC50 obtained using the two electrode voltage clamp assays (column 6) are shown for capsaicin, compounds 1, 2, 3 and Resiniferatoxin (RTX). Values are expressed as average ± standard error (SE).
