Figure 1.
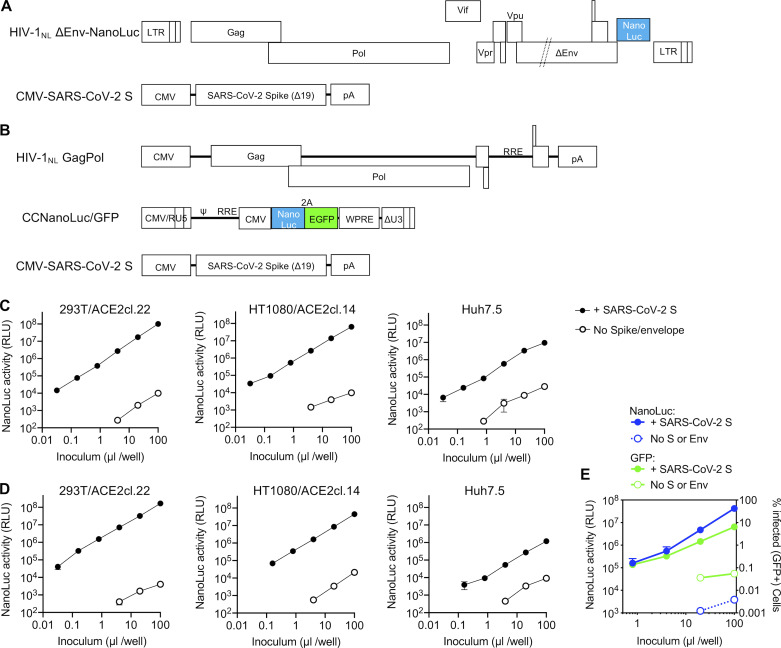
Two-plasmid and three-plasmid HIV-1–based pseudotyped viruses. (A) Schematic representation of the modified HIV-1NL ΔEnv-NanoLuc genome in which a deletion in env was introduced and Nef-coding sequences were replaced by those encoding a NanoLuc luciferase reporter. Infectious virus particles were generated by cotransfection of pHIV-1NL4ΔEnv-NanoLuc and a plasmid encoding the SARS-CoV-2 S lacking the 19 amino acids at the C-terminus of the cytoplasmic tail (SΔ19). (B) Schematic representation of constructs used to generate SARS-CoV-2 S pseudotyped HIV-1–based particles in which HIV-1NLGagPol, an HIV-1 reporter vector (pCCNanoLuc/GFP) encoding both NanoLuc luciferase and EGFP reporter, and the SARS-CoV-2 SΔ19 are each expressed on separate plasmids. RRE, HIV-1 Rev response element; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element. (C) Infectivity measurements of HIV-1NL ΔEnv-NanoLuc particles (generated using the plasmids depicted in A) on the indicated cell lines. Infectivity was quantified by measuring NanoLuc luciferase activity (RLUs) following infection of cells in 96-well plates with the indicated volumes of pseudotyped viruses. The mean and range of two technical replicates are shown. Target cells 293T/ACE2cl.22 and HT1080/ACE2cl.14 are single-cell clones engineered to express human ACE2 (see Fig. S1 A). Virus particles generated in the absence of viral envelope glycoproteins were used as background controls. (D) Same as C, but viruses were generated using the three plasmids depicted in B. (E) Infectivity measurements of CCNanoLuc/GFP containing SARS-CoV-2 pseudotyped particles generated using plasmids depicted in B on 293ACE2*(B) cells, quantified by measuring NanoLuc luciferase activity (RLU) or GFP levels (percentage of GFP-positive cells). Mean and range from two technical replicates are shown.