Figure 3. Design of a cell-free cyanuric acid sensor.
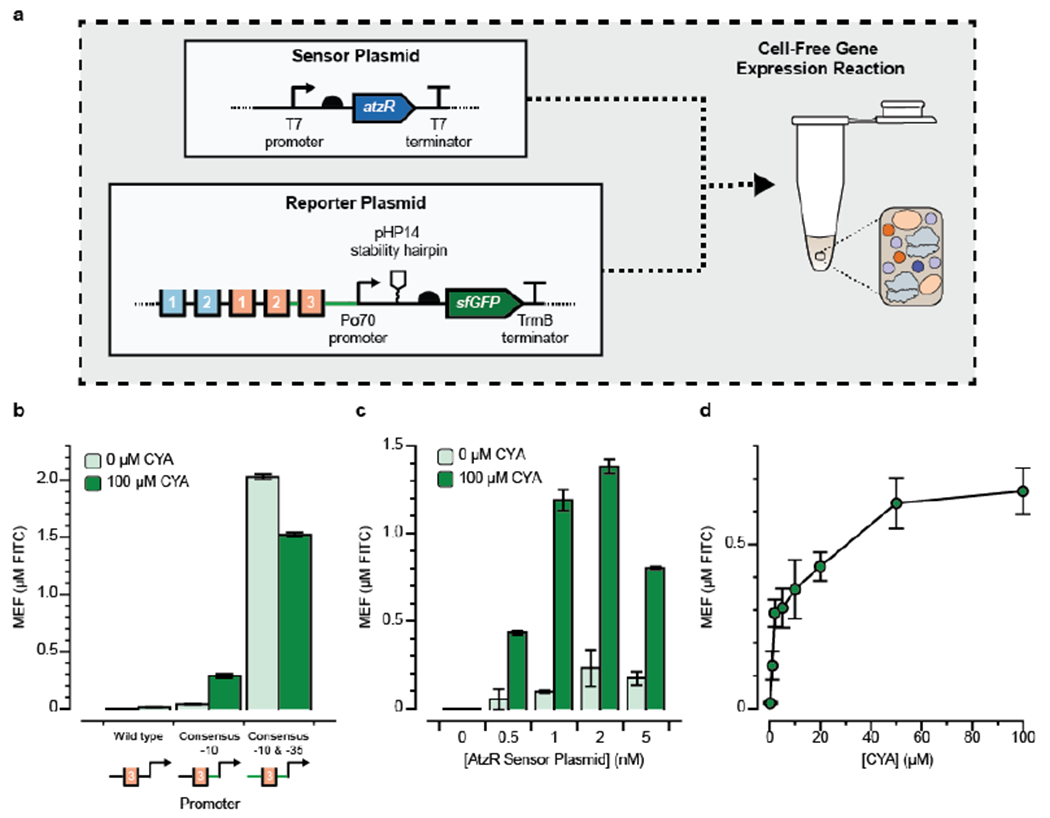
(a) Two plasmids—one encoding the AtzR transcription factor (sensor plasmid) and one encoding sfGFP regulated by AtzR (reporter plasmid)—were added to 10 μL cell-free gene expression reactions and sfGFP production after four hours at 30°C was measured in the absence (OFF) and presence (ON) of 100 μM cyanuric acid (CYA). Plasmid designs are annotated as in Fig. 1. (b) Three promoter variants that replaced the native Pseudomonas σ70 recognition sites with the consensus E. coli −10 and −35 sites were tested in a small screen to identify promoter variants that were functional for cell-free CYA-induced activation. A promoter where only the −10 site was converted to the E. coli consensus sequence (TATAAT) gave the highest fold-induction and was carried through for subsequence experiments. In every case, reporter DNA plasmid was added at 10 nM, and the AtzR-expressing plasmid was kept at 1 nM. (c) Titration of AtzR-expressing plasmid against 10 nM reporter plasmid reveals that the optimal condition has 1 nM of AtzR-expressing plasmid to minimize sensor leak and potentially to maintain adequate distribution of translational resources between the two proteins. (d) Dose-response curve for cyanuric acid induction at optimal plasmid concentrations shows a wide operating range over 0-100 μM CYA. Kinetic plots showing the fluorescence trajectories for all samples in b-d are in Supp. Fig. 3. Error bars represent 1 standard deviation from 3 technical replicates. Fluorescence is reported using mean equivalent fluorescence (MEF) of fluorescein isothiocyanate based on a previously developed standard curve.
