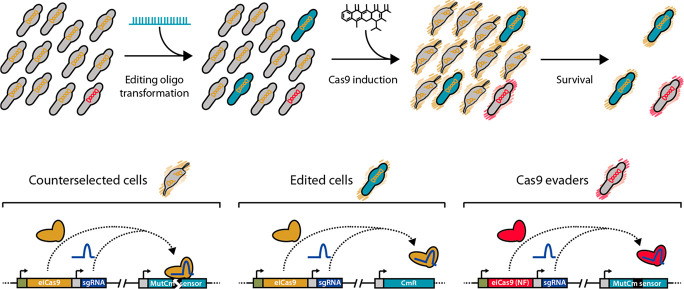
Figure 4.
Outline of the recombineering and Cas9-mediated counterselection strategy. Top: scheme depicting a bacterial population along the different steps of the protocol. The initial population is composed of unedited cells (gray bacteria) carrying a Cas9 system that is either functional (indicated by orange chromosomes) or nonfunctional (Cas9 evaders, indicated by red chromosomes). Upon oligo transformation, some cells become edited (green cells), and subsequent Cas9 induction (represented by orange or red outer shades) results in the selective killing of unedited cells (gray bacteria with broken chromosomes) and the survival of edited cells and Cas9 evaders. Bottom: scheme depicting the molecular mechanisms of this selection. In counterselected cells the functional Cas9 protein (orange molecule) forms a complex with eNT2 sgRNA (purple molecule) that specifically cuts the frameshifting sequence (black box) found in the MutCm sensor. The same complex is formed in edited cells, but the oligo-mediated deletion of the frameshifting sequence precludes Cas9-mediated cleavage of the chromosome. In Cas9 evaders, a nonfunctional (NF) copy of Cas9 is expressed (red molecule), resulting in the survival of cells still carrying the frameshifting sequence on their chromosomes.