Abstract
Depression is a seriously disabling psychiatric disorder with a significant burden of disease. Metabolic abnormalities have been widely reported in depressed patients and animal models. However, there are few systematic efforts that integrate meaningful biological insights from these studies. Herein, available metabolic knowledge in the context of depression was integrated to provide a systematic and panoramic view of metabolic characterization. After screening more than 10 000 citations from five electronic literature databases and five metabolomics databases, we manually curated 5675 metabolite entries from 464 studies, including human, rat, mouse and non-human primate, to develop a new metabolite-disease association database, called MENDA (http://menda.cqmu.edu.cn:8080/index.php). The standardized data extraction process was used for data collection, a multi-faceted annotation scheme was developed, and a user-friendly search engine and web interface were integrated for database access. To facilitate data analysis and interpretation based on MENDA, we also proposed a systematic analytical framework, including data integration and biological function analysis. Case studies were provided that identified the consistently altered metabolites using the vote-counting method, and that captured the underlying molecular mechanism using pathway and network analyses. Collectively, we provided a comprehensive curation of metabolic characterization in depression. Our model of a specific psychiatry disorder may be replicated to study other complex diseases.
Keywords: depression, metabolite, database, pathway analysis, network analysis
Introduction
Depression is a seriously disabling psychiatric disorder with a lifetime prevalence of 20% [1], characterized by high disease burden and excess mortality [2]. From 1990 to 2016, depression was the fifth leading cause of years lived with disability worldwide, contributing 34.1 million of total years lived with disability [3]. As a complex mental illness, the pathogenic factors and clinical manifestations of depression are diverse, and there is obvious heterogeneity among different subtypes [4]. Although a variety of theories of depression have been proposed, the molecular mechanism remains poorly understood, and its treatment faces the challenge that most first-line antidepressants target brain monoamine modulation, which are crude compared with the ideal rapid-acting agents [5].
As the final product of the molecular interactions of genes, transcripts and proteins, metabolites play an important biological role in the organism [6]. With the rapid progress in systems biology over recent decades, metabolomics technologies, including nuclear magnetic resonance, gas chromatography-mass spectrometry and liquid chromatography-mass spectrometry, have been widely applied to identify metabolic characterization in depression and following antidepressant exposure in plasma [7, 8], urine [9] and cerebrospinal fluid [10]. Findings from in vivo magnetic resonance spectroscopy studies have also shown neurometabolite abnormalities in various brain regions of depression patients [11, 12]. Further, evidence from animal models supports an association of metabolite abnormalities in the brain and peripheral tissues with depression and antidepressant exposure [13–15]. However, despite this progress, there are only a few systematic efforts dedicated to integrating the known knowledge from these varied studies [16], and it remains unclear which metabolites are associated with depression.
With the increasing output of high-throughput platforms, knowledge bases for diseases including MetSigDis [17] and DisGeNET [18] have been recently created to gather and display useful molecular information. However, the amount of knowledge data for a specific disease is limited in these pan-disease bases, and the simple annotation scheme still does not address the research needs of complex diseases, including depression. A comprehensive analysis based on large-scale data would provide higher statistical efficiency and more credible biological insights than individual studies. However, few studies have investigated the potential methods of data integration and interpretation across studies for a metabolic database.
Thus, the aim of the present study was to provide a panoramic and systematic view of metabolic characterization in the context of depression by developing a knowledge base of metabolic characterization in depression. To this end, we manually integrated available knowledge for depressed patients and animal models, as well as metabolic changes resulting from treatments, in a new metabolite-disease association database called the metabolite network of depression database (MENDA; http://menda.cqmu.edu.cn:8080/index.php). The standardized data extraction process was used for data collection, a multi-faceted annotation scheme was developed and a user-friendly search engine and web interface were integrated for database access. To facilitate data analysis and interpretation based on MENDA, we also proposed a systematic analytical framework. Case studies were provided that identified the consistently altered metabolites using the vote-counting method, and which captured the underlying molecular mechanism using pathway and network analyses.
Methods
Data collection and curation
Figure 1A illustrates the schematic architecture of the MENDA. Researchers were trained using pilot tests before each step, then two researchers completed data collection and curation independently. All data were checked to identify disagreements, and regular meetings were arranged to resolve any misunderstandings or disagreements.
Figure 1.
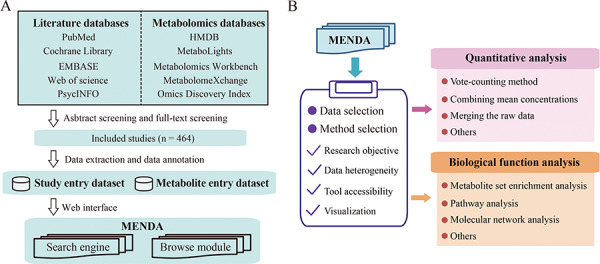
Schematic architecture of the (A) MENDA and the (B) proposed systematic framework for data analysis.
Literature search and study selection
Studies that investigated the metabolic characterization associated with depression, and that used nuclear magnetic resonance, mass spectrometry or magnetic resonance spectroscopy technologies, were collected as follows. Five electronic literature databases (PubMed, Cochrane Library, Embase, Web of Science, and PsycINFO) were searched using the search terms provided in Supplementary Table S1. Five metabolomics databases (Human Metabolome Database (HMDB) [19], MetaboLights [20], Metabolomics Workbench [21], MetabolomeXchange [22] and Omics Discovery Index [23]) were searched with relevant keywords, such as depression, depressive and mood disorder. Further relevant studies were obtained by screening reference lists of all included studies and relevant reviews. The citation lists were also screened in Google Scholar. A total of 11 747 citations from literature databases and 208 citations from metabolomics databases were identified as of 20 March 2018 (Supplementary Table S2).
Retrieved literature citations were imported into Endnote X8 software (Clarivate Analytics; Philadelphia, PA, USA) to remove duplicate records, and the remaining titles and abstracts were then manually reviewed. Using the data inclusion criteria described in the Supplementary Note, a total of 1525 potentially eligible articles were chosen. From these articles, 1064 articles were excluded after the full-text articles were reviewed (Supplementary Table S3), resulting in 464 studies were presented in MENDA.
Data extraction process
Data of interest were manually extracted from the original reports using standardized data abstraction spreadsheets (Supplementary Table S4). Candidate metabolites were selected if the metabolites (including the ratio of two metabolites, e.g. kynurenine/tryptophan ratio) were reported to be significantly changed in the original reports. We chose these criteria because data processing varied across different studies, and significant metabolites were indicated by a P < 0.05 in most of included studies. These criteria were also widely used in other knowledge bases [17, 24].
Data annotation
A dataset containing the study entries (the study entry dataset) and another dataset containing the metabolite entries (the metabolite entry dataset) were generated based on the extracted data. For dataset reformatting and cross-data set mapping, a multi-faceted annotation scheme (Supplementary Table S5) was developed to homogeneously annotate extracted data in both datasets. To create the metabolite entry dataset, metabolite names in the original reports were manually matched in HMDB [19], Kyoto Encyclopedia of Genes and Genomes (KEGG) [25] and PubChem [26] for the purpose of metabolite name standardization, then each metabolite entry was annotated with external source identifiers and study-related information using the annotation scheme.
Database architecture
A multilayer relational database was created for data storage and management utilizing the MySQL 5.5 database system (https://www.mysql.com/). A user-friendly search engine and web interface were incorporated for users to search and browse depression-associated metabolic alterations and relevant studies. The Perl script was used for Common Gateway Interface programming. For web browsing, the front page of the retrieval system was based on HTML. The Apache HTTP Server 2.4 (http://httpd.apache.org/) was used as the web server of the retrieval system.
Framework for comprehensive analysis of metabolic characterization
To facilitate data analysis and interpretation for users, we proposed a systematic framework (Figure 1B). Users can download the dataset provided in MENDA and perform personalized analysis. Users choose the appropriate methods based on research objectives, data heterogeneity, tool accessibility and graphic output. In this study, we performed the following analyses for providing case studies. The details for data selection are provided in the Supplementary Note.
Data integration
The vote-counting method was used to identify the consistently up-regulated or down-regulated metabolites across the combined studies, and the numbers of up-regulated and down-regulated differential metabolites in the original reports were counted. A binomial test was then used to evaluate whether a given metabolite was consistently regulated across the combined studies, with the assumption of a probability of 0.5 that a given metabolite is upregulated in each study. The one-tailed P-value was calculated using the function ‘binom.test’ in R software version 3.4.4 (https://www.r-project.org/). Statistical significance was set at a Benjamini–Hochberg procedure adjusted false discovery rate (FDR) < 0.05. Only metabolites reported for least six different datasets were selected for analysis.
Biological function analysis
Details of the analysis are provided in the Supplementary Note. In brief, metabolite set enrichment analysis and metabolic pathway analysis were performed using MetaboAnalyst 4.0 [27] to identify the significantly disturbed metabolite sets and metabolic pathways, respectively. Canonical pathway analysis and molecular network analysis were then performed using Ingenuity Pathway Analysis (IPA; http://www.ingenuity.com). Statistical significance was set at an FDR < 0.05 in all analyses.
Results
Data statistics in MENDA
The detail of data statistics in MENDA are shown in Figure 2, and the information for each included study and each metabolite entry (the study entry dataset and metabolite entry dataset) are provided in Supplementary Data: MENDA.xlsx. From 464 included studies, we collected 5675 differential metabolite entries. Most studies were conducted to identify candidate metabolites between depressed and healthy states (type 1 studies, N = 391; with 3206 metabolite entries), or to identify candidate metabolites resulting from treatments in the depressed state (type 2 studies, N = 151; with 1402 metabolite entries). Figure 3 summarizes the numbers of metabolite entries for the most frequently reported metabolites in separate settings. Tryptophan metabolism-related metabolites (serotonin, 5-hydroxyindoleacetic acid, quinolinic acid and tryptophan) were the most frequently changed metabolites after treatment, which may be explained by the monoamine modulation effects of current antidepressants [28].
Figure 2.
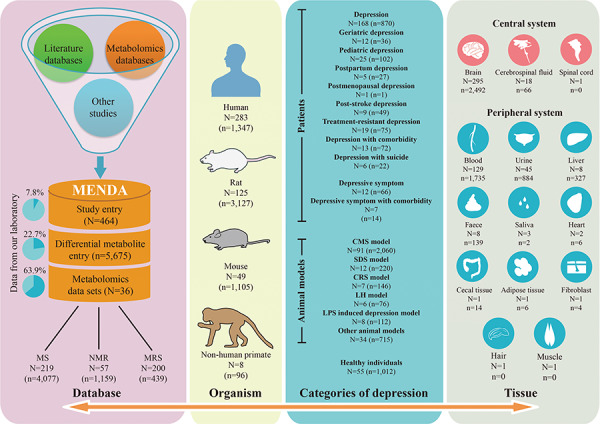
Data statistics of MENDA. N, number of studies; n, number of differential metabolite entries; CMS, chronic mild stress; SDS, social defeat stress; CRS, chronic restraint stress; LH, learned helplessness; LPS, lipopolysaccharide; MRS, magnetic resonance spectroscopy; MS, mass spectrometry; NMR, nuclear magnetic resonance.
Figure 3.
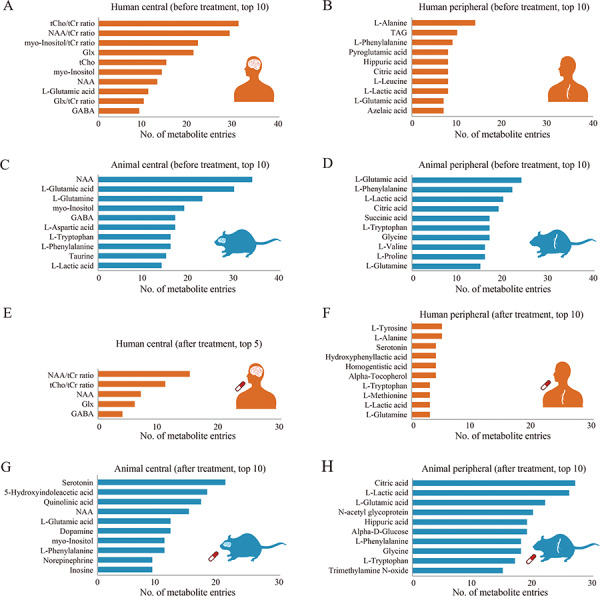
The numbers of entries for the most frequently reported metabolites in separate settings. The plots summarize the numbers of entries for the most frequently reported metabolites in the central (A) and peripheral (B) systems of patients, in the central (C) and peripheral (D) systems of animal models before treatment, and in the central (E) and peripheral (F) systems of patients, and the central (G) and peripheral (H) systems of animal models, after treatment. Orange and blue bars denote the numbers of metabolite entries from human and animal models for the specific metabolites, respectively. GABA, gamma-aminobutyric acid; Glx, glutamate and glutamine; NAA, N-acetyl-L-aspartic acid; tCho, choline-containing compounds; tCr, creatine and phosphocreatine.
The distribution of metabolite entries in 18 tissues is shown in Supplementary Figure S1. In brief, in type 1 studies that compared the metabolic characterization between depressed and healthy states, 20 metabolites in patients and 92 in animal models were reported as differential metabolites in at least three tissues (Supplementary Figure S1A). In type 2 studies that identified differential metabolites resulting for depression, 8 metabolites in patients and 52 in animal models were reported as differential metabolites in at least three tissues (Supplementary Figure S1B).
Web interfaces in MENDA
A user-friendly web interface and search engine were incorporated for researchers in MENDA (http://menda.cqmu.edu.cn:8080/index.php), as described below.
Data browsing
In the browse section, four entry points were offered. (i) General: the systematic reviews of metabolic characterization for human, rodent, non-human primate and all organisms were provided (Supplementary Figure S2A). In each detailed page, relevant studies and metabolites were displayed based on the subcategories of depression. (ii) Metabolite: all metabolites collected in MENDA were listed alphabetically (Supplementary Figure S2B). The metabolite name, external source identifiers (HMDB, KEGG and PubChem IDs), synonyms and relevant studies were displayed in detailed pages. (iii) Study: the included studies in MENDA were listed numerically (Supplementary Figure S2C). Relevant information (including title, overall design, study type, data available, organism, categories of depression, criteria for depression, sample size, tissue, platform, paper links and differential metabolites) for each study was provided in detailed pages. (iv) Metabolite map: a graphical network generated by the function ‘networkD3’ in R was presented to show the relationships between tissues (blue nodes) and relevant metabolites (yellow nodes).
Data search
In the search section, both quick search and advanced search approaches were provided (Supplementary Figure S2D). (i) Quick search: a quick search by metabolite names, external source identifiers and fuzzy words was provided. (ii) Advanced search: five search options (study type, tissue type, organism, category of depression and platform) were incorporated for data filtering. Relevant studies and metabolites with the hyperlinks were shown in the Search Result page.
Others
The study entry dataset, the metabolite entry dataset, and 23 metabolomics datasets from our previous published studies were available as Excel files, and the hyperlinks of 13 datasets from other research teams were listed. Data are free for download to all users. Other information for this database, such as data statistics and tutorial, was provided in other sections. Data updating and database structure upgrading in MENDA is an ongoing process.
Application of MENDA for data integration
The vote-counting method was used to identify which metabolites were consistently altered. Among the 202 metabolites that were introduced to the vote-counting process in all settings, 18 metabolites were consistently up-regulated across studies, and 24 were consistently down-regulated (Supplementary Table S6). For example, depressed patients had lower levels of brain gamma-aminobutyric acid and glutamate/glutamine (Figure 4A), which is consistent with the findings of previous meta-analyses [29, 30].
Figure 4.
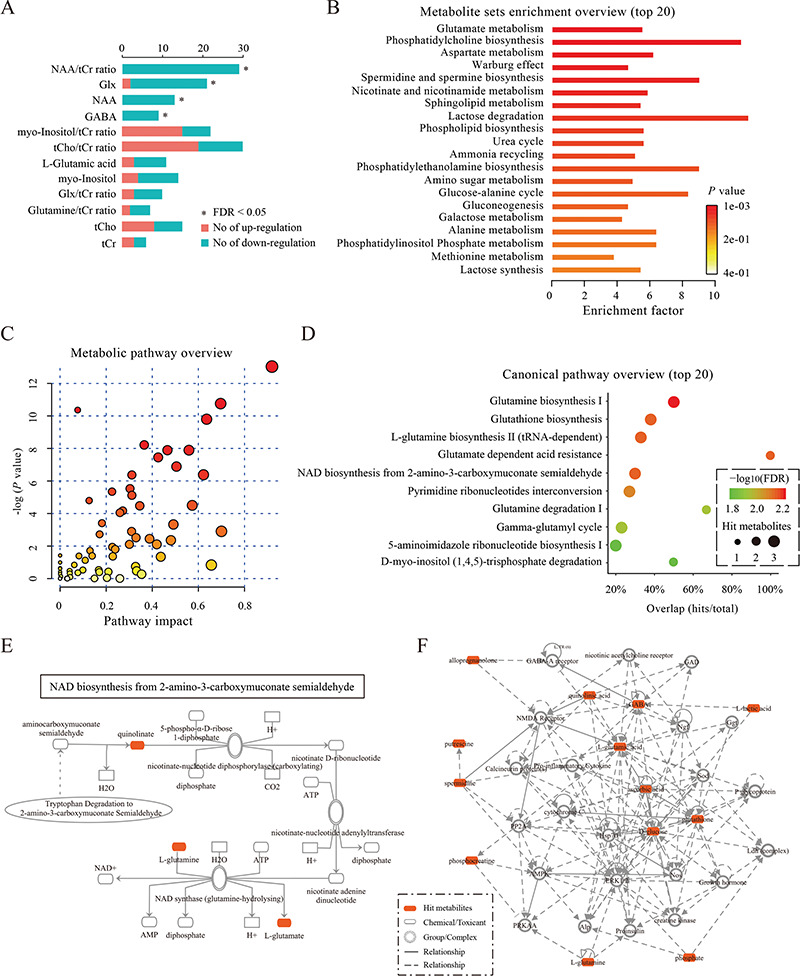
Plots for data integration and biological function analysis. (A) The plot for the results of the vote-counting method. The plot summarizes the distribution of up-regulated or down-regulated of metabolites across studies. Red and blue bars denote the numbers of studies that reported that the metabolite was up-regulated or down-regulated, respectively. An asterisk (*) indicates an FDR < 0.05. (B) The plot for the results of metabolite set enrichment analysis. The top 20 enriched metabolite sets are shown. For each metabolite set, the color of the bars denotes the P-value of the hypergeometric test, and the ‘enrichment factor’ was calculated by dividing the number of uploaded metabolites (hits) by the expected number of matches. (C) The plot for the results of metabolic pathway analysis. Nodes represent metabolic pathways, the x-axis shows the −log10(P-value), and the y-axis shows the pathway impact. (D) The plot for the results of canonical pathway analysis. Nodes represent metabolic pathways, the x-axis shows the pathway names, and the y-axis shows the overlap rate of numbers actually matched from the user-uploaded metabolites (hits) and the total number of molecules in the pathways (total). (E) The plot for a canonical pathway. Metabolites, proteins and the interrelation in this canonical pathway are presented. (F) The plot for a molecular network. Metabolites, proteins and the interrelation in this network are presented.
Application of MENDA for biological function analysis
We also explored the enriched metabolite sets and disturbed metabolic pathways using MetaboAnalyst. Enriched metabolite sets across separate settings are shown in Supplementary Table S7. For example, 19 metabolite sets in the central nervous system of patients were enriched with P < 0.05, while none were significantly enriched with an FDR < 0.05 (Figure 4B). The results of the metabolic pathways analysis showed that 14 metabolic pathways were significantly altered before treatment, and 19 pathways after treatment (Figure 4C and Supplementary Table S8). Interestingly, most of these pathways were shared both before and after treatment. To provide more detailed information on the differential metabolites involved in each pathway associated with depression, we integrated the 14 significantly disturbed metabolic pathways into a simplified pathway diagram (Figure 5). Another pathway diagram is also provided for the seven metabolic pathways with P < 0.05 but an FDR > 0.05 (Supplementary Figure S3).
Figure 5.
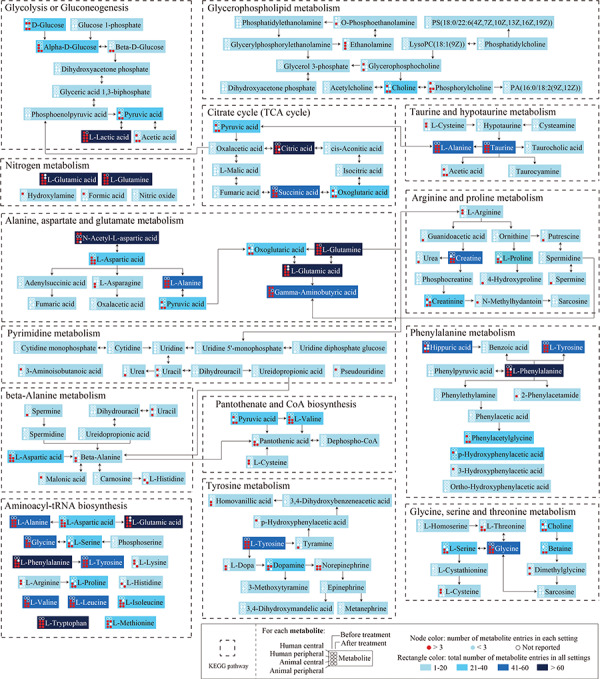
Metabolites and interactions in the 14 significantly altered metabolic pathways. Dotted boxes represent the significantly altered KEGG pathways (metabolic pathway analysis FDR < 0.05). Rectangles represent the metabolites in the pathways. For each metabolite, nodes in each row represent the number of metabolite entries in the human central, human peripheral, animal central and animal peripheral systems, respectively; nodes in the first and second columns represent the numbers of metabolite entries before and after treatment, respectively. Red node: number of metabolite entries ≥3; blue node: number of metabolite entries <3; hollow node: number of metabolite entries = 0. Colored rectangles represent the total number of metabolite entries across all eight settings, with darker colors representing larger numbers.
We then identified the significantly altered canonical pathways (Supplementary Table S9) and disturbed molecular networks (Supplementary Table S10) using IPA. For example, 41 canonical pathways in the human central nervous system were significantly enriched before treatment (Figure 4D). An example of a canonical pathway is shown in Figure 4E. ‘Cellular compromise, lipid metabolism, small molecule biochemistry’ was the only significantly disturbed molecular network in the human central nervous system, with a score of 35 (Figure 4F). Canonical pathway and network analyses also showed an important role of the glutamate system in depression, which supports previous reports that glutamate receptors are potential targets for the development of novel antidepressant agents [31–33]. One quarter of the top 20 canonical pathways in the central nervous system were shared between patients and animal models, and 55% in the peripheral system were shared between patients and animal models. These findings implicated that animal models still could not mimic all the molecular changes of patients, and more studies based on human participants are needed to confirm the findings from animal models.
Discussion
There is increasing evidence for metabolite abnormalities in brain and peripheral tissues in depression and with antidepressant treatment (Supplementary Figure S4). To provide a full view of current knowledge from a perspective of systems biology, we manually developed a new metabolite-disease association database. We also proposed a framework for big-data driven data integration and biological function analysis, which can provide insights for the curated knowledge of MENDA. To our knowledge, this is the first metabolic database for a specific neuropsychiatry disease. Our model of a single psychiatry disorder may also be replicated to study other complex diseases.
The aim of the present study was to manually collect and annotate all available metabolic knowledge of depression from the literature and other databases. To achieve this goal, we manually curated 5675 metabolite entries from 464 studies through systematic searches in 10 databases and full-text screening from thousands articles. Compared with the MetSigDis that only contains 37 metabolite entries for depression [17], we provided a hundred-fold increase in metabolite entries. Nevertheless, more disease-specific metabolic knowledge bases are required to address the biochemical research needs of complex diseases. The use of a standardized data extraction process and multi-faceted annotation scheme also enabled us to present an overview of metabolic characterization, which may be a valuable resource for researchers interested in depression or database development.
In addition to data presentation in MENDA, we proposed a systematic framework of data integration and biological function analysis to clarify underlying biological information from heterogeneous data sources. For data integration, the vote-counting method was chosen to combine data, as raw metabolic datasets or mean concentrations of metabolites were not accessible in many studies [34]. This method has been used in previous large-scale systems biology studies [35, 36]. Other statistical methods, including combining mean concentrations [37, 38] and merging the raw data [27], are potential choices for specific datasets from studies that have provided the mean concentrations or even raw metabolomics data.
Compared with traditional methods that only focus on a small number of metabolites, biological function analysis that integrates complex information from heterogeneous datasets allows for analysis of all available candidate molecules within a systematic framework, to elucidate the biological mechanism in complex diseases [39, 40]. Pathway and network analyses, which examine the interactions between metabolites, genes and proteins within biological pathways or networks [41, 42], are the most common methods for big-data driven research [43, 44]. In addition to MetaboAnalyst or IPA mentioned above, users can choose other potential tools for bioinformatics analysis.
There are two major limitations of this study. First, like other knowledge bases [45, 46], candidate metabolites were collected based on the statistical threshold in the original reports. Further statistical correction and bioinformatics analysis are needed when the full quantitative data are available from many studies. However, the number of available metabolome datasets in MENDA remains limited, as many obstacles remain for the goal of making data findable, accessible, interoperable and reusable [47]. Second, all the biological processes of depression cannot be understood solely by metabolic changes [48]. Thus, we will integrate other omics data, including proteomics and genomics, in future research.
Conclusion
After screening more than 10 000 citations, we manually curated 5675 metabolite entries from 464 studies in MENDA (http://menda.cqmu.edu.cn:8080/index.php). The standardized data extraction process and the multi-faceted annotation scheme enabled us to systematically provide a panoramic view of metabolic characterization in depression. A user-friendly search engine and web interface were integrated for database access. To facilitate data analysis and interpretation based on MENDA, we also proposed a systematic analytical framework, including data integration and biological function analysis. Our model, which systematically integrates metabolic knowledge base construction for a specific psychiatry disorder, may be replicated to study other complex diseases.
Key Points
MENDA is the first metabolic database for a specific neuropsychiatry disease.
A multi-faceted annotation scheme containing study-level and metabolite-level knowledge and a user-friendly interface were provided in MENDA, which may serve as an important resource for researchers interested in depression or database development.
MENDA presents a panoramic view of metabolic characterization in depression that is based on manual curation of 5675 differential metabolite entries from 464 studies.
Based on MENDA, we proposed a systematic analytical framework, including data integration and biological function analysis, to facilitate data analysis and interpretation.
Case studies were provided for the systematic analytical framework.
Supplementary Material
Acknowledgements
We thank Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Juncai Pu is a PhD candidate in the Department of Neurology, The First Affiliated Hospital of Chongqing Medical University.
Yue Yu is a bioinformatics post-doctoral fellow in the College of Medical Informatics, Chongqing Medical University, and in the Department of Health Sciences Research, Mayo Clinic.
Yiyun Liu is a PhD candidate in the Department of Neurology, The First Affiliated Hospital of Chongqing Medical University.
Lu Tian is a postgraduate student in the Institute of Neuroscience, Chongqing Medical University.
Siwen Gui is a PhD candidate in the Institute of Neuroscience, Chongqing Medical University.
Xiaogang Zhong is a postgraduate student in the Institute of Neuroscience, Chongqing Medical University.
Chu Fan is a postgraduate student in the College of Medical Informatics, Chongqing Medical University.
Shaohua Xu is a postgraduate student in the Institute of Neuroscience, Chongqing Medical University.
Xuemian Song is a postgraduate student in the Institute of Neuroscience, Chongqing Medical University.
Lanxiang Liu is a PhD candidate in the Department of Neurology, The First Affiliated Hospital of Chongqing Medical University.
Lining Yang is a PhD candidate in the Department of Neurology, The First Affiliated Hospital of Chongqing Medical University.
Peng Zheng is an associate professor in the Department of Neurology, The First Affiliated Hospital of Chongqing Medical University.
Jianjun Chen is a staff scientist in the Institute of Neuroscience, Chongqing Medical University.
Ke Cheng is a neuroscience post-doctoral fellow in the Institute of Neuroscience, Chongqing Medical University.
Chanjuan Zhou is an associate professor in the Institute of Neuroscience, Chongqing Medical University.
Haiyang Wang is a PhD candidate in the Institute of Neuroscience, Chongqing Medical University.
Peng Xie is a professor and the director of the Institute of Neuroscience, Chongqing Medical University.
Funding
This work was funded by the National Key Research and Development Program of China (grant number 2017YFA0505700).
References
- 1. Hasin DS, Sarvet AL, Meyers JL, et al. . Epidemiology of adult DSM-5 major depressive disorder and its specifiers in the United States. JAMA Psychiat 2018;75:336–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cuijpers P, Vogelzangs N, Twisk J, et al. . Comprehensive meta-analysis of excess mortality in depression in the general community versus patients with specific illnesses. Am J Psychiatry 2014;171:453–62. [DOI] [PubMed] [Google Scholar]
- 3. Vos T, Abajobir AA, Abate KH, et al. . Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the global burden of disease study 2016. Lancet 2017;390:1211–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goldberg D. The heterogeneity of ‘major depression’. World Psychiatry 2011;10:226–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harmer CJ, Duman RS, Cowen PJ. How do antidepressants work? New perspectives for refining future treatment approaches. Lancet Psychiatry 2017;4:409–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patti GJ, Yanes O, Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol 2012;13:263–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu X, Zheng P, Zhao X, et al. . Discovery and validation of plasma biomarkers for major depressive disorder classification based on liquid chromatography-mass spectrometry. J Proteome Res 2015;14:2322–30. [DOI] [PubMed] [Google Scholar]
- 8. Ji Y, Hebbring S, Zhu H, et al. . Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: pharmacometabolomics-informed pharmacogenomics. Clin Pharmacol Ther 2011;89:97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng P, Wang Y, Chen L, et al. . Identification and validation of urinary metabolite biomarkers for major depressive disorder. Mol Cell Proteomics 2013;12:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Erhardt S, Lim CK, Linderholm KR, et al. . Connecting inflammation with glutamate agonism in suicidality. Neuropsychopharmacology 2013;38:743–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gabbay V, Hess DA, Liu S, et al. . Lateralized caudate metabolic abnormalities in adolescent major depressive disorder: a proton MR spectroscopy study. Am J Psychiatry 2007;164:1881–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Merkl A, Schubert F, Quante A, et al. . Abnormal cingulate and prefrontal cortical neurochemistry in major depression after electroconvulsive therapy. Biol Psychiatry 2011;69:772–9. [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y, Yuan S, Pu J, et al. . Integrated metabolomics and proteomics analysis of hippocampus in a rat model of depression. Neuroscience 2018;371:207–20. [DOI] [PubMed] [Google Scholar]
- 14. Li B, Guo K, Zeng L, et al. . Metabolite identification in fecal microbiota transplantation mouse livers and combined proteomics with chronic unpredictive mild stress mouse livers. Transl Psychiatry 2018;8:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Webhofer C, Gormanns P, Tolstikov V, et al. . Metabolite profiling of antidepressant drug action reveals novel drug targets beyond monoamine elevation. Transl Psychiatry 2011;1:e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tenenbaum JD, Bhuvaneshwar K, Gagliardi JP, et al. . Translational bioinformatics in mental health: open access data sources and computational biomarker discovery. Brief Bioinform 2017. doi: 10.1093/bib/bbx157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng L, Yang H, Zhao H, et al. . MetSigDis: a manually curated resource for the metabolic signatures of diseases. Brief Bioinform 2019;20:203–9. [DOI] [PubMed] [Google Scholar]
- 18. Piñero J, Bravo À, Queralt-Rosinach N, et al. . DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 2017;45:D833–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wishart DS, Feunang YD, Marcu A, et al. . HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 2018;46:D608–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haug K, Salek RM, Conesa P, et al. . MetaboLights - an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res 2013;41:D781–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sud M, Fahy E, Cotter D, et al. . Metabolomics workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res 2016;44:D463–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leiden University MetabolomeXchange http://www.metabolomexchange.org (17 September 2018, date last accessed).
- 23. Perez-Riverol Y, Bai M, da Veiga Leprevost F, et al. . Discovering and linking public omics data sets using the Omics discovery index. Nat Biotechnol 2017;35:406–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang L, Chang S, Li Z, et al. . ADHDgene: a genetic database for attention deficit hyperactivity disorder. Nucleic Acids Res 2012;40:D1003–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanehisa M, Furumichi M, Tanabe M, et al. . KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y, Xiao J, Suzek TO, et al. . PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res 2009;37:W623–W633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chong J, Soufan O, Li C, et al. . MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 2018;46:W486–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mann JJ. The medical management of depression. N Engl J Med 2005;353:1819–34. [DOI] [PubMed] [Google Scholar]
- 29. Schür RR, Draisma LW, Wijnen JP, et al. . Brain GABA levels across psychiatric disorders: a systematic literature review and meta-analysis of (1) H-MRS studies. Hum Brain Mapp 2016;37:3337–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taylor MJ. Could glutamate spectroscopy differentiate bipolar depression from unipolar? J Affect Disord 2014;167:80–4. [DOI] [PubMed] [Google Scholar]
- 31. Zarate CA, Jr, Mathews D, Ibrahim L, et al. . A randomized trial of a low-trapping nonselective N-methyl-D-aspartate channel blocker in major depression. Biol Psychiatry 2013;74:257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maeng S, Zarate CA, Jr, Du J, et al. . Cellular mechanisms underlying the antidepressant effects of ketamine: role of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptors. Biol Psychiatry 2008;63:349–52. [DOI] [PubMed] [Google Scholar]
- 33. Shin S, Kwon O, Kang JI, et al. . mGluR5 in the nucleus accumbens is critical for promoting resilience to chronic stress. Nat Neurosci 2015;18:1017–24. [DOI] [PubMed] [Google Scholar]
- 34. Goveia J, Pircher A, Conradi LC, et al. . Meta-analysis of clinical metabolic profiling studies in cancer: challenges and opportunities. EMBO Mol Med 2016;8:1134–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rhodes DR, Yu J, Shanker K, et al. . Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A 2004;101:9309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nilsson R, Jain M, Madhusudhan N, et al. . Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun 2014;5:3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guasch-Ferré M, Hruby A, Toledo E, et al. . Metabolomics in prediabetes and diabetes: a systematic review and meta-analysis. Diabetes Care 2016;39:833–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park JE, Lim HR, Kim JW, et al. . Metabolite changes in risk of type 2 diabetes mellitus in cohort studies: a systematic review and meta-analysis. Diabetes Res Clin Pract 2018;140:216–27. [DOI] [PubMed] [Google Scholar]
- 39. Lee WY, Bachtiar M, Choo CCS, et al. . Comprehensive review of hepatitis B virus-associated hepatocellular carcinoma research through text mining and big data analytics. Biol Rev Camb Philos Soc 2019;94:353–367. [DOI] [PubMed] [Google Scholar]
- 40. Hu Y, Pan Z, Hu Y, et al. . Network and pathway-based analyses of genes associated with Parkinson's disease. Mol Neurobiol 2017;54:4452–65. [DOI] [PubMed] [Google Scholar]
- 41. Mitra K, Carvunis AR, Ramesh SK, et al. . Integrative approaches for finding modular structure in biological networks. Nat Rev Genet 2013;14:719–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shin SY, Fauman EB, Petersen AK, et al. . An atlas of genetic influences on human blood metabolites. Nat Genet 2014;46:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hu YS, Xin J, Hu Y, et al. . Analyzing the genes related to Alzheimer's disease via a network and pathway-based approach. Alzheimers Res Ther 2017;9:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Monti C, Colugnat I, Lopiano L, et al. . Network analysis identifies disease-specific pathways for Parkinson's disease. Mol Neurobiol 2018;55:370–81. [DOI] [PubMed] [Google Scholar]
- 45. Gutiérrez-Sacristán A, Grosdidier S, Valverde O, et al. . PsyGeNET: a knowledge platform on psychiatric disorders and their genes. Bioinformatics 2015;31:3075–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chang SH, Gao L, Li Z, et al. . BDgene: a genetic database for bipolar disorder and its overlap with schizophrenia and major depressive disorder. Biol Psychiatry 2013;74:727–33. [DOI] [PubMed] [Google Scholar]
- 47. Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. . The FAIR guiding principles for scientific data management and stewardship. Sci Data 2016;3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cambiaghi A, Ferrario M, Masseroli M. Analysis of metabolomic data: tools, current strategies and future challenges for omics data integration. Brief Bioinform 2017;18:498–510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


