Figure 5. A unifying genetic model of ASD and its connection with the ASD molecular perturbations.
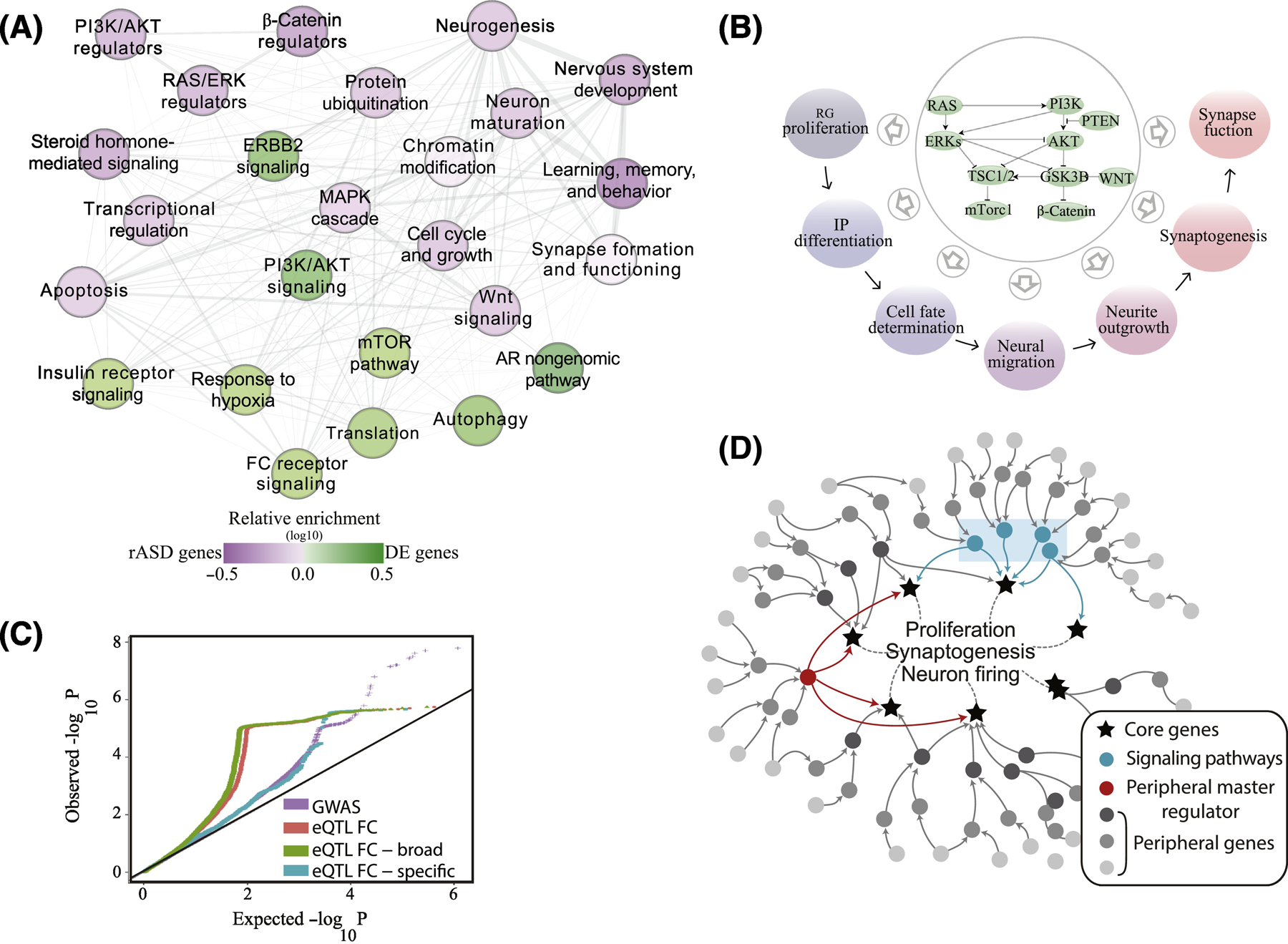
(A) In a recent study of large samples of ASD and typical toddlers, analyses of leukocyte gene expression identified a highly interconnected network of differentially expressed genes in ASD. In the panel (from[51]), each node represents a biological process that is significantly enriched in an expanded ASD network that included the differentially expressed genes and ASD risk genes. Nodes in green are signaling pathways (eg., PI3K/AKT, RAS/ERK, mTOR) and prenatal processes that are perturbed transcriptionally in leukocytes from toddlers with ASD. The purple nodes are enriched with ASD risk genes including risk ASD regulator genes that target the signaling pathways. Thus, many ASD risk genes and common variants are not directly involved in core prenatal processes underpinning ASD, but instead dysregulate such processes in ASD at critical prenatal and early postnatal time points that are relevant to ASD. Fig. 2E shows this same differentially expressed ASD gene network displays high gene coexpression across brain regions during 1st and 2nd trimester of development. (B) The PI3K/AKT, RAS/ERK, WNT/β-catenin signaling pathways that are dysregulated in leukocytes of ASD toddlers and correlate with their social symptom severity, are, in fact, highly conserved and their signaling can modulate multiple, Epoch-1 and Epoch-2 neurodevelopmental processes. (C) ASD liability resides in SNPs with broadly-expressed regulatory genes with roles across multiple tissues. GWAS data were retrieved from Grove et al[32]. Frontal cortex (FC) eQTLs were retrieved from the GTEx portal[64]. A frontal cortex eQTL was deemed as broadly functional if it was significantly correlated with gene expression levels in ≥50% of diverse tissues[64]. A similar pattern was observed from available GWAS data on schizophrenia[65]. (D) Illustration of proposed genetic architecture of ASD where regulatory ASD genes and common variants are not directly involved in core prenatal processes underpinning ASD, but instead are upstream in highly interconnected “peripheral” regulatory networks (dark and medium gray dots) that regulate downstream core brain genes (stars) and processes (eg., proliferation, synaptogenesis) at critical prenatal and early postnatal time points relevant to ASD[51]. Peripheral network dysregulations due to mutations in these regulatory ASD genes may propagate through gene regulatory networks to disrupt transcriptional programs or signaling pathways (blue rectangle) which impact downstream core brain-specific genes and processes related to ASD. Master regulators, such as CHD8, are proximal to and regulate directly key brain genes and processes and, so, their mutation can have dramatic impact on them.
