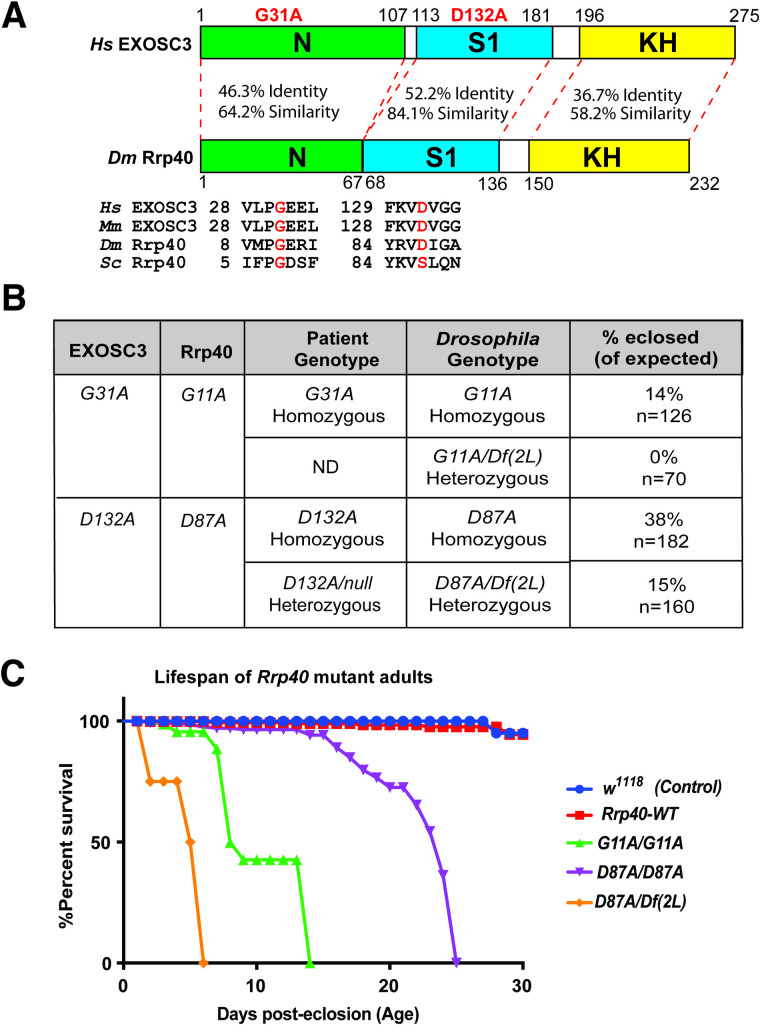
Fig 3. Generation of Drosophila models of PCH1b amino acid substitutions.
(A) The EXOSC3/Rrp40 protein consists of three domains: an N-terminal domain (N), a central S1 putative RNA binding domain, and a C-terminal putative RNA binding KH (K homology) domain. The disease-causing amino acid substitutions (G31A and D132A) modeled in Drosophila in this study are indicated in red above the domain structure. The position and flanking sequence of the amino acid substitutions in PCH1b-associated human EXOSC3 (shown in red) and the corresponding amino acids in Mus musculus (Mm), Drosophila melanogaster (Dm), and Saccharomyces cerevisiae (Sc) are shown. The percent identity/similarity between the human and Drosophila domains is indicated. (B) The amino acid substitutions that occur in EXOSC3 in patients that are modeled in Drosophila Rrp40, are indicated with Patient Genotype (Homozygous/Heterozygous) as modeled in Drosophila (Drosophila Genotype) shown. For each Drosophila model, the viability of that genotype engineered in flies by CRISPR/Cas9 gene editing is shown as % eclosed (of expected). Rrp40 mutants show decreased viability, indicated by skewed Mendelian ratios. n = the number of individual flies analyzed. The G11A over null genotype has not been reported in patients-indicated as not detected (ND) under patient genotype. (C) A Kaplan-Meier analysis shows that Rrp40 mutant flies with the indicated genotypes to model patient genotypes exhibit early adult lethality compared to wildtype Control flies (w1118) or Rrp40-WT (Rrp40wt) flies that have undergone CRISPR/Cas9 gene editing to produce a control wildtype Rrp40 allele (see Materials and methods).