Abstract
Long-chain non-coding RNA (lncRNA) Myosin Heavy Chain Associated RNA Transcripts (MHRT) are newly identified cardioprotective lncRNAs. In this study, we investigated the association of MHRT gene single nucleotide polymorphisms with risk and prognosis of chronic heart failure (CHF).
Sanger sequencing was performed to detect the genotypes of rs3729830, rs7140721, rs76614781, rs3729829, rs3729828, rs3729825, and rs3729822 loci in the non-coding region of the MHRT gene from 240 patients with CHF and 240 control subjects. After 3 years of follow-up, progression-free survival was recorded in patients with CHF.
The risk of CHF in subjects carrying A allele of the MHRT gene rs7140721 locus was 1.43 times higher than that of C allele carriers (95% CI: 1.23–1.62, P < .001). The risk of CHF in subjects carrying A allele of the rs3729829 locus was 1.41 times higher than that of G allele carriers (95% CI: 1.20–1.61, P < .01). The risk of CHF in the carriers of T allele of the rs3729825 locus was 1.89 times higher than that of C allele carriers (adjusted OR = 1.89, 95% CI: 1.66–2.04, P < .01). Further, the level of lncRNA MHRT in the plasma of subjects carrying CA/AA genotype of the rs7140721 locus was significantly higher than that of subjects carrying the CC genotype. The level of lncRNA MHRT in the plasma of subjects carrying GA/AA genotype of the rs3729829 locus was significantly higher than that of subjects carrying the GG genotype. In addition, the level of lncRNA MHRT in subjects with CT/TT genotype of the rs3729825 locus carriers was significantly higher than that in subjects with the CC genotype (P < .05). In addition, significant differences in the mortality of patients with CHF were observed between different genotypes of rs7140721, rs3729829, and rs3729825 loci (P < .001).
The single nucleotide polymorphisms of MHRT gene rs7140721, rs3729829, and rs3729825 loci were associated with the risk of CHF and prognosis.
Keywords: chronic heart failure, long-chain non-coding RNA, myosin heavy chain associated RNA transcripts, single nucleotide polymorphism
1. Introduction
Chronic heart failure (CHF) is a common complication of cardiovascular disease, the incidence of which is increasing yearly, and is a serious threat to people's health and life.[1] Early diagnosis and evaluation are essential for rational treatment and reduction of mortality in patients with CHF; however, there are few biomarkers for effective early diagnosis and prognostic assessment.[2]
Long-chain non-coding RNAs (lncRNAs) are a class of functional RNAs, over 200 nt in length, located in the nucleus or cytoplasm, and are universally transcribed in eukaryotic cells, but have little or no function in encoding proteins.[3] Recent studies have shown that lncRNAs are involved in the occurrence and development of many diseases in humans, such as cancer[4] and coronary artery disease.[5] There is evidence showing that lncRNAs are also associated with the occurrence and development of CHF. For example, Yu et al[6] found that lncRNA UCA1 contributes to the diagnosis and prognosis of CHF. The plasma level of lncRNA UCA1 is likely to be a good indicator for the diagnosis of CHF and to predict a poor prognosis for CHF.
lncRNA Myosin Heavy Chain Associated RNA Transcripts (MHRT) are newly identified cardioprotective lncRNAs whose transcripts have been shown to be a novel ATP-dependent chromatin remodeling factor lncRNA for heart failure.[7] In a cardiomyocyte injury model induced by H2O2 in neonatal rats, the expression of lncRNA MHRT was elevated, and si-RNA MHRT increased cell apoptosis induced by H2O2.[7] In addition, the MHRT/Nrf2 pathway is involved in the regulation of doxorubicin-induced apoptosis in cardiomyocytes.
In the present study, we selected several single nucleotide polymorphisms (SNP) loci with minor allele frequencies (MAF) > 0.05 in the non-coding region of the MHRT gene, rs3729830, rs7140721, rs76614781, rs3729829, rs3729828, rs3729825, and rs3729822, and explored the correlation of these SNPs with the risk and prognosis of CHF in the Chinese Han population.
2. Materials and methods
2.1. Subjects
We randomly selected 240 patients with CHF who underwent treatments in the Second Affiliated Hospital of Zhejiang Chinese Medical University and the Hangzhou Hospital of Traditional Chinese Medicine between February 2014 and February 2016, including 125 males and 115 females. Their age ranged from 45 to 89 years and the average age was 61.66 ± 10.34 years. There were 186 cases of ischemic cardiomyopathy and 54 cases of dilated cardiomyopathy. All patients with heart failure were aged between 40 and 85 years and had chronic and stable symptoms of heart failure.[8] Patients were contacted by the outpatient service or telephone interviews, and a 3-year follow-up was conducted for cardiovascular mortality.
The criteria for recruitment were:
-
(1)
meet the diagnostic criteria of CHF, or have left ventricular enlargement or reduction in left ventricular ejection fraction;
-
(2)
have a basic disease that can clearly result in CHF; and
-
(3)
aged 18 years or older, and undergone heart failure treatment within 1 or more than 1 month from the start of the study.
The exclusion criteria were:
-
(1)
patients with malignant tumors or severe infectious diseases, history of organ transplantation, and acute decompensated heart failure;
-
(2)
patients with occurrence of acute coronary events within 1 month prior to the start of the study, such as unstable angina, acute ST elevation myocardial infarction, and acute non-ST elevation myocardial infarction; and
-
(3)
serum creatinine > 2.5 mg/mL.
A cohort of 240 non-CHF subjects was recruited at the physical examination center of the same hospital as a control group and excluded organic heart disease and other acute and chronic systematic diseases, abnormal electrocardiogram, and excluded abnormal cardiac structure and cardiac functions by echocardiography. The recruitment procedure was in accordance with the principles of the World Medical Association Declaration of Helsinki with all participants signing informed consent forms, and this study was approved by the Medical Ethics Committee of the Second Affiliated Hospital of Zhejiang Chinese Medical University and the Hangzhou Hospital of Traditional Chinese Medicine.
2.2. Genotype analysis of the SNPs
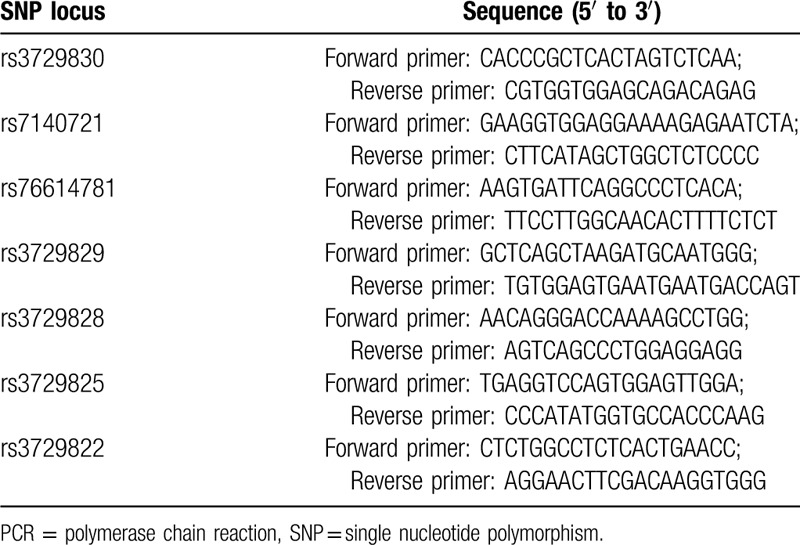
According to the Variation View (https://www.ncbi.nlm.nih.gov/variation/view/) database, we selected 7 SNPs which were located in introns of the MHRT gene with MAF > 0.05, rs3729830, rs7140721, rs76614781, rs3729829, rs3729828, rs3729825, and rs3729822. Genomic DNA was extracted from the plasma of participants using a Puregene DNA Isolation Kit (Qiagen, Minneapolis, MN, USA) according to the manufacturer's instructions, and a DNA fragment containing the SNP locus was obtained by PCR amplification. The primer sequences of each SNP locus are shown in Table 1. The primers for PCR were synthesized by Genewiz (Suzhou, Shanghai). The PCR was conducted in a 25 μL system containing 50 ng extracted genomic DNA, 0.5 μL 10 mM dNTP, 2.5 μL 10 × PCR buffer, 1.5 μL 25 mM Mg2+, 0.25 μL 5 U/μL Taq, and 1 μL 10 pmol of forward and reverse primers. The PCR conditions were: 95 °C, 5 min; 94 °C, 45 s; 60 °C, 45 s; 72 °C, 30 s, 35 cycles; and a final extension at 72 °C for 10 minutes. The genotype of the PCR products was determined using Sanger sequencing, and 20% of the samples were genotyped blindly and the same procedure was used to obtain the results.
Table 1.
PCR primer sequences for SNPs.
2.3. Real-time quantitative PCR (qRT-PCR)
Total RNA was extracted from the plasma of participants using the TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Then, the extracted RNA was reverse transcribed into cDNA using PrimeScript RT Master Mix kit (TaKaRa, Dalian, China). According to the manufacturer's instructions, qRT-PCR was performed using the PrimeScript RT kit and SYBR Premix Ex Taq (TaKaRa, Dalian, China). The primers for lncRNA MHRT qRT-PCR were: 5′-ACACGGCGTTCTTGAGTTT-3′ (forward); 5′-AGTATGAGGAGTCGCAGTCG-3′ (reverse). GAPDH was used as an internal reference and the primers were: 5′-GTCAACGGATTTGGTCTGTATT-3′ (forward); 5′-AGTCTTCTGGGTGGCAGTGAT-3′ (reverse). PCR conditions were: 95 °C, 30 seconds; 95 °C, 5 seconds; and 60 °C, 34 seconds for 40 cycles. The qRT-PCR was performed using an ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA), and the fold change in expression of lncRNA MHRT relative to GAPDH was calculated using the 2-ΔΔCT method.
2.4. Statistical analyses
Statistical analyses were conducted using SPSS (version 22.0, SPSS Inc., Chicago, IL). Continuous variables are expressed as mean ± SD and statistical analyses were performed using an independent-sample t-test. Categorical variables are expressed as n (%) and statistical analyses were performed using a Chi-square test. A Chi-square test was performed to detect the Hardy-Weinberg equilibrium of the MHRT gene. The association between MHRT gene SNPs and the risk of CHF was estimated by an unconditional logistic regression analysis, adjusted to factors such as age, gender, hypertension, and diabetes. The Kaplan-Meier curve and the Cox proportional hazard regression model were used to estimate the effect of MHRT gene SNPs on the survival of patients with CHF. All studies were 2-sided, with P < .05 considered statistically significant.
3. Results
3.1. Comparison of clinical characteristics
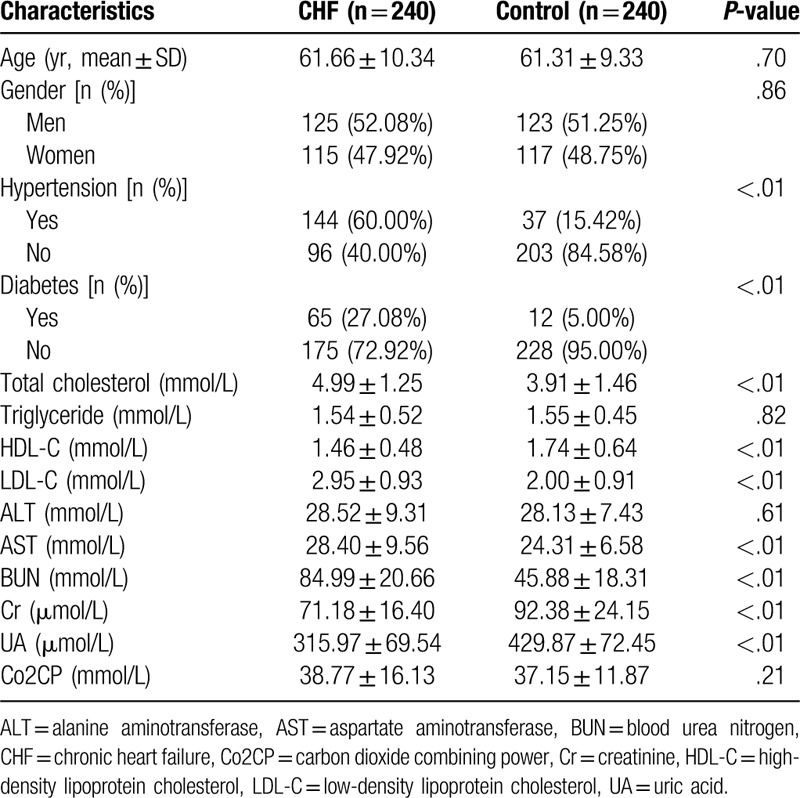
The general characteristics of the 240 patients with CHF and the 240 healthy participants are shown in Table 2. Statistical analyses showed that there was no significant difference in age, gender, plasma triglyceride level, ALT level, and Co2CP level between the 2 groups (P > .05). The proportion of patients with hypertension and patients with diabetes in the CHF group was higher than that in the control group (P < .05). The levels of total cholesterol, LDL-C, AST, and BUN were significantly higher than those of the control group, while the levels of HDL-C, Cr and UA were significantly lower than those of the control group (P < .05).
Table 2.
Comparison of clinical characteristics between patients with CHF and the control group.
3.2. Correlation between SNPs of the MHRT gene and the risk of CHF occurrence
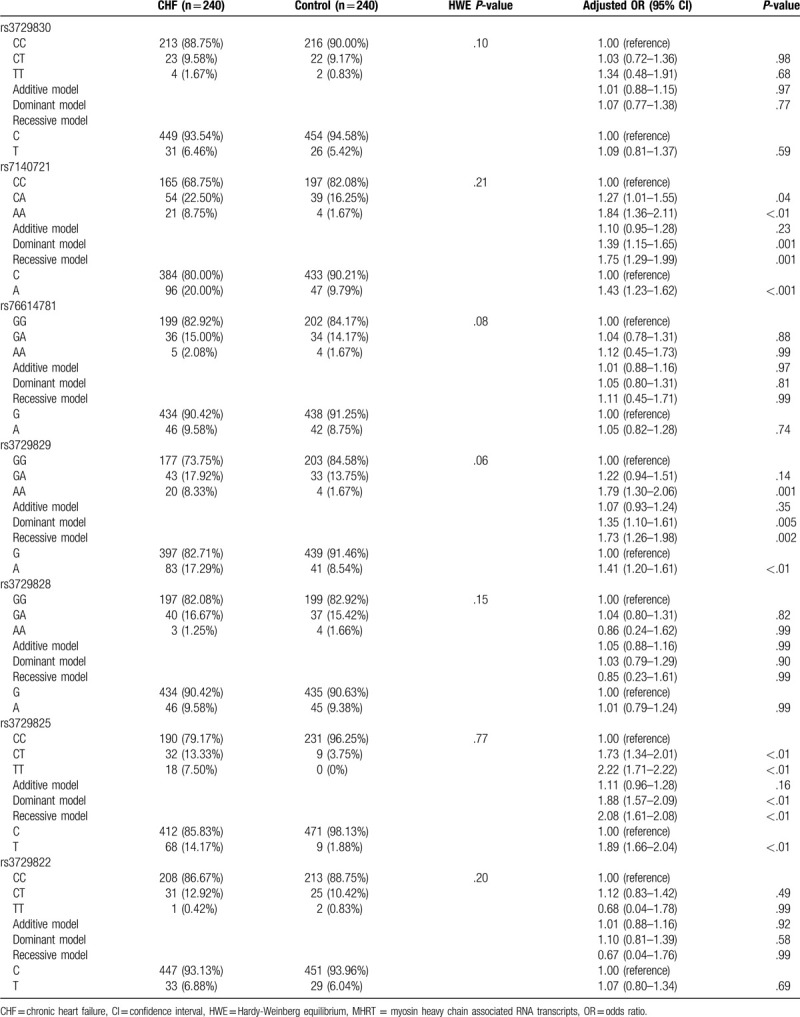
The genotype frequencies of the MHRT gene rs3729830, rs7140721, rs76614781, rs3729829, rs3729828, rs3729825, and rs3729822 loci were in accordance with the Hardy-Weinberg equilibrium (HWE) (P > .05) (Table 3). We found that there were no significant differences in the frequency of genotypes and alleles of the MHRT gene rs3729830, rs76614781, and rs3729828 loci between patients with CHF and the control group (P > .05). However, taking the CC genotype of the MHRT gene rs7140721 as a reference, the frequencies of CA and AA genotypes in patients with CHF were significantly higher than those in the control group (P < .05), and the risk of CHF occurrence was elevated in both the dominant and recessive models (adjusted OR = 1.39, 95% CI: 1.15–1.65, P = .001; adjusted OR = 1.75, 95% CI: 1.29–1.99, P = .001; respectively). The risk of CHF in the A allele carriers was 1.43 times higher than C allele carriers of the gene (95% CI: 1.23–1.62, P < .001). The risk of CHF occurrence in subjects carrying AA genotype of the rs3729829 locus was 1.79 times higher than the GG genotype carriers (95% CI: 1.30–2.06, P = .001). Although the risk of CHF did not increase in the additive model (adjusted OR = 1.07, 95% CI: 0.93–1.24, P = .35), the risk of CHF was significantly increased in both the dominant and recessive models (adjusted OR = 1.35, 95% CI: 1.10–6.11, P = .005; adjusted OR = 1.73, 95% CI: 1.26–1.98, P = .002; respectively). The risk of CHF in subjects carrying the A allele of the rs3729829 locus was 1.41 times higher than G allele carriers (95% CI: 1.20–1.61, P < .01). Taking the CC genotype of the rs3729825 as a reference, the risk of CHF was significantly increased in CT and TT genotypes (adjusted OR = 1.73, 95% CI: 1.34–2.01, P < .01; adjusted OR = 2.22, 95%CI: 1.71–2.22, P < .01; respectively). There was no significant increase in the risk of CHF in the additive model (adjusted OR = 1.11, 95% CI: 0.96–1.28, P = .16); however, in the dominant and recessive models, the risk of CHF was significantly increased (adjusted OR = 1.88, 95% CI: 1.57–2.09, P < .01; adjusted OR = 2.08, 95% CI: 1.61–2.08, P < .01; respectively), and the risk of CHF in T allele carriers was 1.89 times higher than that of the C allele carriers (adjusted OR = 1.89, 95% CI: 1.66–2.04, P < .01).
Table 3.
Correlation between genotype and allele frequency of MHRT gene SNPs and the risk of CHF.
3.3. Haplotype analysis
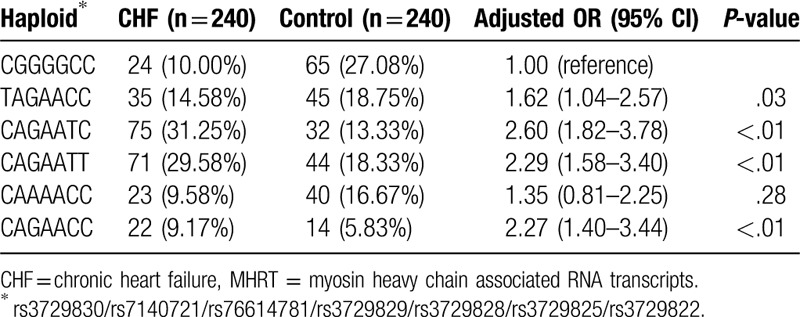
We used Haploview 4.2 to analyze the linkage disequilibrium of MHRT gene rs3729830, rs7140721, rs76614781, rs3729829, rs3729828, rs3729825, and rs3729822 loci (Fig. 1). We found that there were 4 haploids which were associated with an increased risk of CHF: TAGAACC (adjusted OR = 1.62, 95% CI: 1.04–2.57, P = .03), CAGAATC (adjusted OR = 2.60, 95% CI: 1.82–3.78, P < .01), CAGAATT (adjusted OR = 2.29, 95% CI: 1.58–3.40, P < .01), and CAGAACC (adjusted OR = 2.27, 95% CI: 1.40–3.44, P < .01) (Table 4).
Figure 1.
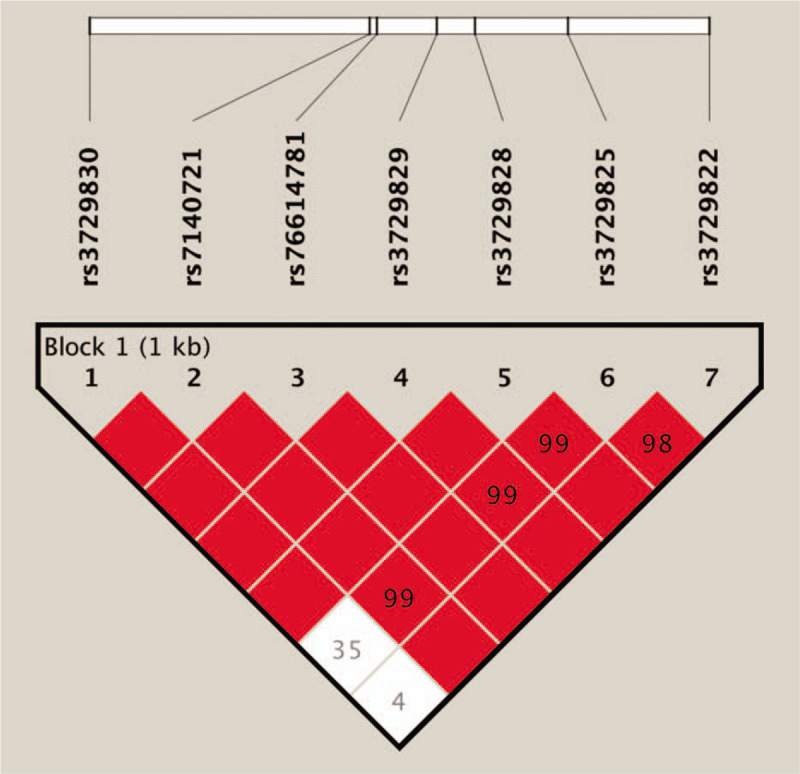
Analysis of linkage disequilibrium of the SNPs of MHRT genes via Haploview 4.2. The number in the box represents D’. The larger the value, the stronger the degree of linkage, and the lack of numbers in the red box represent D’ = 1. MHRT = myosin heavy chain associated RNA transcripts, SNP = single nucleotide polymorphism.
Table 4.
Association of MHRT gene haplotypes with the risk of CHF.
3.4. Stratified analysis
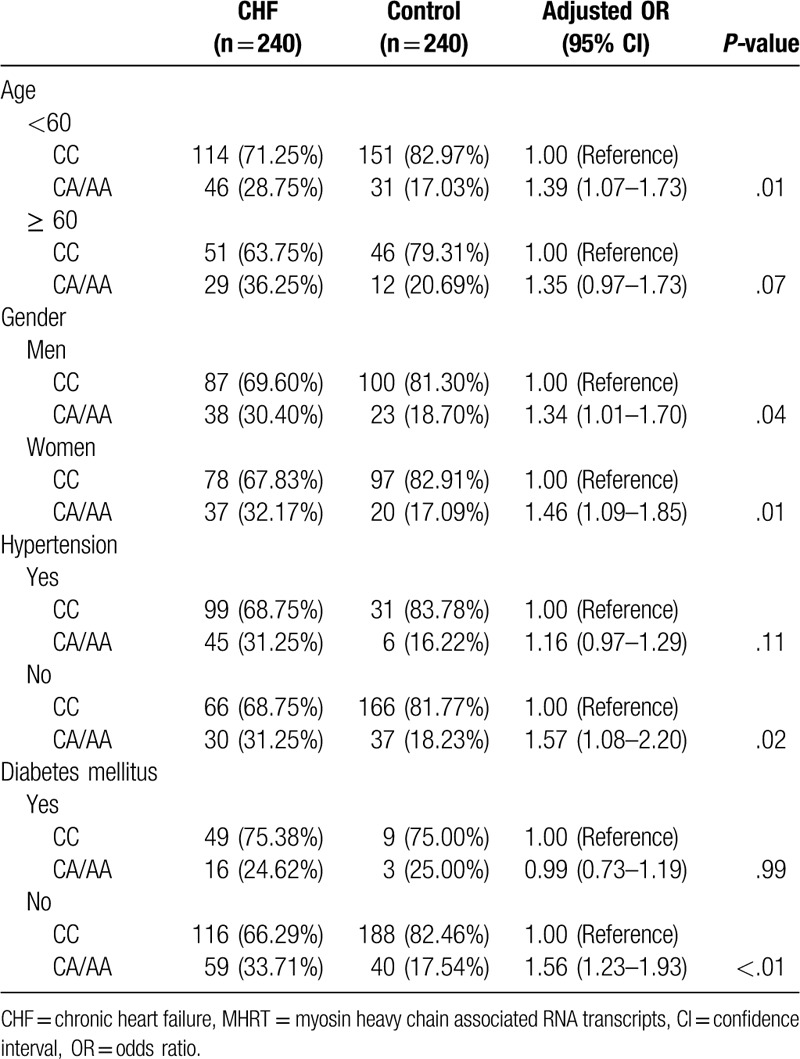
We then conducted a stratified analysis of the general characteristics of the subjects and divided all participants into young subjects (age < 60 years) and elderly subjects (age ≥ 60 years), men and women, patients with and without hypertension and patients with and without diabetes. The results showed that the risk of CHF in the carriers of A allele (CA/AA) of the rs7140721 locus was significantly higher than that of the CC genotype carriers in the young population (adjusted OR = 1.39, 95% CI: 1.07–1.73, P = .01), men and women (adjusted OR = 1.34, 95% CI: 1.01–1.70, P = .04; adjust OR = 1.46, 95% CI: 1.09–1.85, P = .01; respectively), patients without hypertension (adjusted OR = 1.57, 95% CI: 1.08–2.20, P = .02), and patients without diabetes (adjusted OR = 1.56, 95% CI: 1.23–1.93, P < .01). However, there was no alteration in the risk of CHF in the A allele (CA/AA) of the rs7140721 carriers compared to that in the CC genotype carriers in the elderly, patients with hypertension and patients with diabetes (P > .05) (Table 5).
Table 5.
Stratified analysis of the relationship between different genotypes of MHRT gene rs7140721 locus and the risk of CHF.
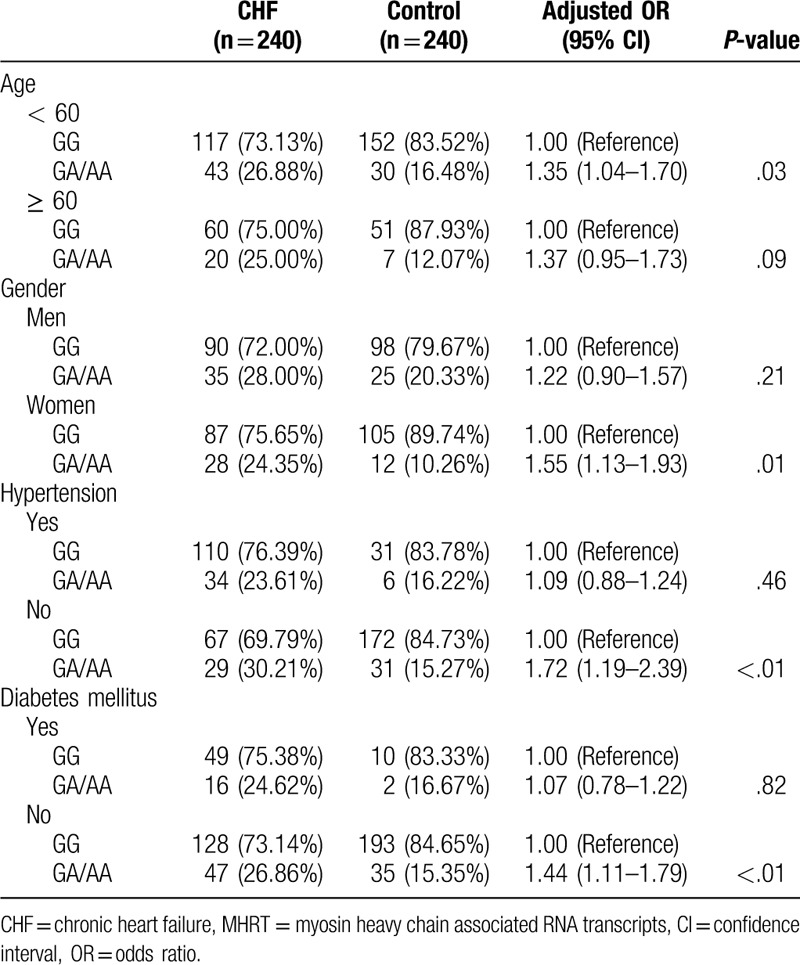
The risk of CHF in the carriers of the A allele (GA/AA) of the rs3729829 locus was significantly higher than that of the GG genotype carriers in the young population (adjusted OR = 1.35, 95% CI: 1.04–1.70, P = .03), women (adjusted OR = 1.55, 95% CI: 1.13–1.93, P = .01), patients without hypertension (adjusted OR = 1.72, 95% CI: 1.19–2.39, P < .01), and patients without diabetes (adjusted OR = 1.44, 95% CI: 1.11–1.79, P < .01) but not in the elderly, male, and patients with hypertension and diabetes (P > .05) (Table 6).
Table 6.
Stratified analysis of the relationship between different genotypes of MHRT gene rs3729829 locus and the risk of CHF.
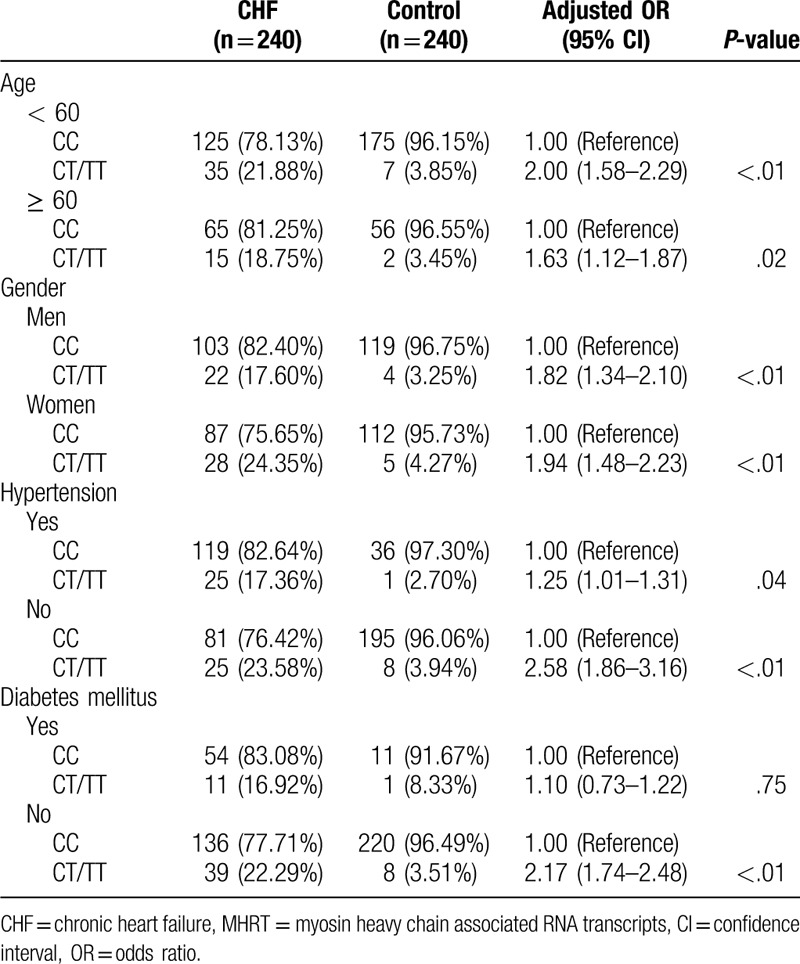
Additionally, only in the patients without diabetes, was the risk of CHF in the rs3729825 T allele carriers (CT/TT) not significantly increased (adjusted OR = 2.17, 95% CI: 1.74–2.48, P < .01) (Table 7).
Table 7.
Stratified analysis of the relationship between different genotypes of MHTR gene rs3729825 locus and the risk of CHF.
3.5. Correlation between plasma level of lncRNA MHRT and MHRT gene SNP
Further, we performed qRT-PCR to analyze the level of lncRNA MHRT in the plasma of the subjects. The results showed that the plasma lncRNA MHRT level in patients with CHF was significantly higher than that in the control group (P < .01) (Fig. 2A). We found that the levels of lncRNA MHRT in the plasma of rs7140721 locus CA/AA genotype carriers were significantly higher than those of CC genotype in both patients with CHF and control subjects (P < .05) (Fig. 2B, 2E). The levels of lncRNA MHRT in the plasma of rs3729829 locus GA/AA genotype carriers were significantly higher than those of GG genotype in both patients with CHF and the control subjects (P < .05) (Fig. 2C, 2F). In addition, the levels of lncRNA MHRT in the plasma of rs3729825 locus CT/TT genotype carriers were significantly higher than those of CC genotype in both patients with CHF and the control subjects (P < .05) (Fig. 2D, 2G).
Figure 2.
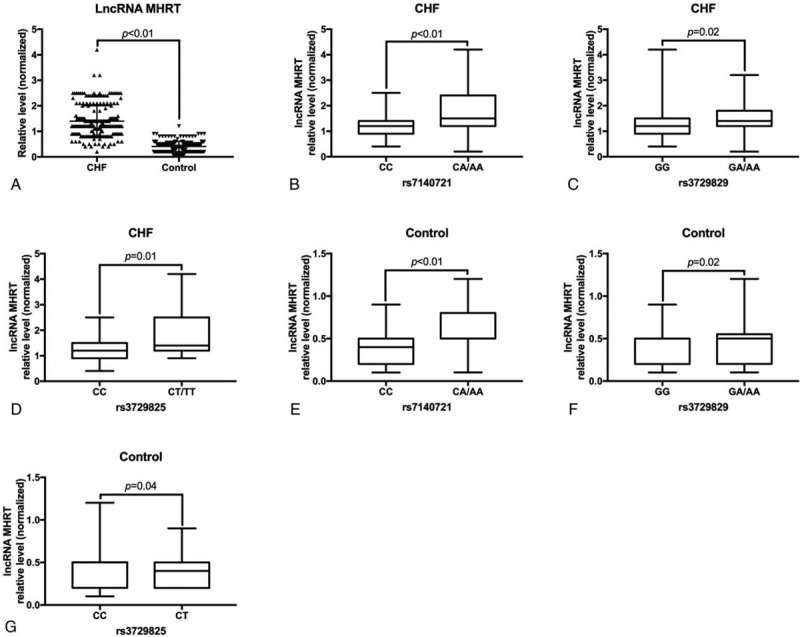
Comparison of plasma lncRNA MHRT levels. (A) Comparison of plasma lncRNA MHRT levels in patients with CHF and control subjects. (B) Comparison of plasma lncRNA MHRT levels in different genotype carriers of rs7140721 locus of patients with CHF. (C) The same as (B) except for rs3729829 locus. (D) The same as (B) except for rs3729825 locus. (E) The same as (B) except for control subjects. (F) The same as (C) except for control subjects. (G) The same as (D) except for control subjects. CHF = chronic heart failure, LncRNA = long-chain non-coding RNA, MHRT = myosin heavy chain associated RNA transcripts.
3.6. Association of MHRT gene polymorphisms with progression-free survival in patients with CHF
A total of 240 patients with CHF were followed up, and the median survival of these patients was 31.5 months. We observed a significant difference in the mortality of patients with CHF between different genotypes of rs7140721, rs3729829, and rs3729825 loci (P < .001) (Fig. 3).
Figure 3.
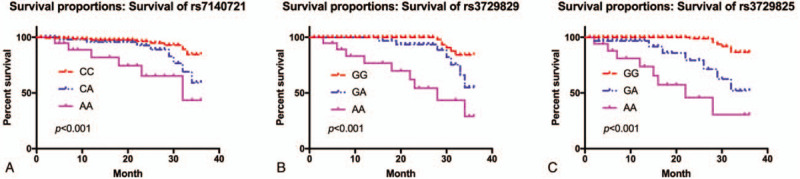
Correlation between MHRT gene polymorphisms and prognosis in patients with CHF. (A) Correlation between different genotypes of rs7140721 locus and prognosis of patients suffering from CHF. (B) The same as (A) except for rs3729829 locus. (C) The same as (A) except for rs3729825 locus. CHF = chronic heart failure, MHRT = myosin heavy chain associated RNA transcripts.
3.7. Correlation of plasma lncRNA MHRT level and progression-free survival (PFS) of patients with CHF
QRT-PCR was used to detect plasma lncRNA MHRT level, we compared between the high-level lncRNA MHRT (above average) and low-level lncRNA MHRT levels (below average) with PFS of patients with CHF. The results showed that patients with higher plasma lncRNA MHRT had lower PFS, and the difference was significant (P = .02, Fig. 4).
Figure 4.
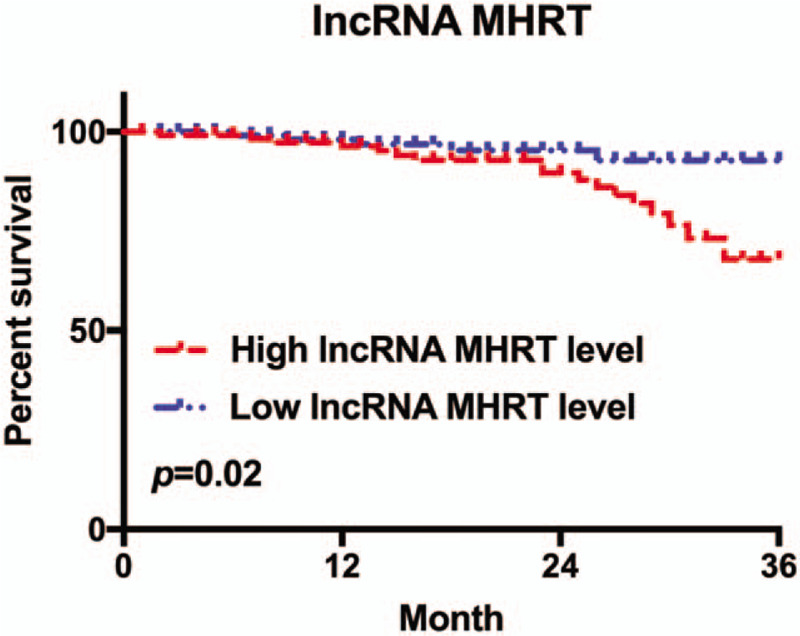
Correlation between plasma lncRNA MHRT level and the PFS of patients with CHF. CHF = chronic heart failure, LncRNA = long-chain non-coding RNA, MHRT = myosin heavy chain associated RNA transcripts.
4. Discussion
In this study, we analyzed the relationship between 7 SNP loci of the MHRT gene non-coding region with MAF > 0.05 and the risk and prognosis of CHF in Chinese Han patients using a case-control study. The results showed that there was a significant correlation between the risk and prognosis of CHF and rs7140721, rs3729829, and rs3729825 SNPs of the MHRT gene.
Recently, cardiovascular diseases have become a major public health problem in China, and the incidence and mortality of cardiovascular diseases are gradually increasing.[9–11] Heart failure is the end stage of various heart diseases. It is a group of pathological syndromes with cardiac insufficiency that seriously affects life and health in humans.[12,13] Hence, early diagnosis and effective treatment are extremely important to reduce the rate of hospitalization and improve the survival rate of patients with heart failure.
lncRNA is a family of RNA with a length of above 200 nt and contributes to various important physiological processes such as cell differentiation, development, defensive response, and immune response.[14,15] Although lncRNA does not encode a protein, it can participate in post-transcriptional regulation and miRNA regulation via epigenetic and transcriptional pathways.[16–18]MHRT is located on chromosome 14q11.2, which encodes an lncRNA that acts as a cardioprotective factor in the heart.[19,20] Previous studies of the similar gene in mice found that its encoding transcripts can regulate chromatin remodeling by acting as a bait for the BRG1 chromatin inhibitor complex and prevent its binding to its genomic target. Blocking BRG1 is essential for protecting the heart from pathological hypertrophy.[7] Additionally, evidence showed that elevation of circulating lncRNA predicts HF and can be considered as a novel biomarker for HF.[21]
Here, we found that the expression level of lncRNA MHRT in plasma of patients with CHF was significantly higher than that of the control group, indicating that the expression level of lncRNA MHRT in plasma of patients with CHF was abnormally increased, which was consistent with the results reported by Xuan et al.[21] We analyzed the correlation between the MHRT gene SNPs and the risk of CHF, and found that only SNPs of rs7140721, rs3729829, and rs3729825 loci were associated with the risk of CHF. The risk of CHF in the A allele carriers of MHRT gene rs7140721 locus was 1.43 times that of C allele carriers. The rs3729829 A allele carriers were 1.41 times more likely to have CHF than the G allele carriers. The risk of CHF in the s3729825 T allele carriers was 1.99 times higher than that of C allele carriers, while other SNP loci were not risk factors for CHF. Considering this result, it is likely that genotypes of the MHRT gene rs7140721, rs3729829, and rs3729825 loci predict the risk of CHF in the earlier stages and it is necessary to take relevant precautions to prevent the occurrence of CHF.
To further analyze the association between the risk of CHF and MHRT gene SNPs in different subpopulations, we performed stratified analyses to investigate the effects of age, gender, hypertension, and diabetes. We found that in young people but not the elderly or patients without hypertension, and diabetes, the risk of CHF in the rs7140721 locus A allele (CA/AA) carriers of the MHRT gene was significantly higher than that of the CC genotype carriers, suggesting the importance of screening for the genotype of MHRT gene rs7140721 locus in these subpopulations. In addition, the risk of CHF in the rs3729829 locus allele (GA/AA) carriers of the MHRT gene in the young and female patients, and patients without hypertension and diabetes was significantly higher than that of GG genotype carriers, indicating that genotype detection of MHRT gene rs3729829 in such subpopulations may be more clinically significant than in the elderly and male patients, and patients with hypertension and diabetes. Moreover, the risk of CHF in the MHRT gene T allele carriers (CT/TT) was not significantly increased only in patients without diabetes, indicating genotyping of the MHRT gene rs3729825 locus in patients without diabetes was not as important as in the other subpopulations.
In this study, all patients were followed up for 3 years. The results showed that the A allele of MHRT gene rs7140721 and rs3729829 loci, and T allele of rs3729825 locus carriers had a poor prognosis; more clinical attention should be paid to these subpopulations.
There are some limitations in the present study. First, the conclusions of this study need to be further verified in a large sample to balance the errors caused by small samples. Second, the specific mechanism of MHRT in the process of CHF has not been demonstrated in this study and further studies are needed to clarify the underlying mechanism. In addition, this study only analyzed the correlation between MHRT and CHF; there may be many other lncRNAs involved in the occurrence of CHF, which requires further screening.
5. Conclusion
In summary, the lncRNA MHRT gene non-coding region rs7140721, rs3729829, and rs3729825 loci SNPs are correlated with the risk and prognosis of CHF and the mechanisms may be related to expression levels of lncRNA MHRT; however, further studies are needed to confirm the underlying mechanism.
Author contributions
G.Z. and Y.C. conceived and designed the experiments. G.Z. and L.D. performed the experiments. G.Z. and L.D. analyzed the data. Y.C. wrote the paper.
Footnotes
Abbreviations: CHF = chronic heart failure, LncRNA = long-chain non-coding RNA, MAF = minor allele frequencies, MHRT = myosin heavy chain associated RNA Transcripts, qRT-PCR = real-time quantitative PCR, SNPs = single nucleotide polymorphisms.
How to cite this article: Zhang G, Dou L, Chen Y. Association of long-chain non-coding RNA myosin heavy chain associated RNA gene single nucleotide polymorphism with risk and prognosis of chronic heart failure. Medicine. 2020;99:29(e19703).
This work was supported by grants from the Traditional Chinese Medical science and technology plan of Zhejiang Province (2018ZA052).
The authors have no funding and conflicts of interest to disclose.
References
- [1].Abuzaanona A, Lanfear D. Pharmacogenomics of the natriuretic peptide system in heart failure. Curr Heart Fail Rep 2017;14:536–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fu TC, Lin YC, Chang CM, et al. Validation of a new simple scale to measure symptoms in heart failure from traditional Chinese medicine view: a cross-sectional questionnaire study. BMC Complement Altern Med 2016;16:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Busch A, Eken SM, Maegdefessel L. Prospective and therapeutic screening value of non-coding RNA as biomarkers in cardiovascular disease. Ann Transl Med 2016;4:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res 2017;77:3965–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hu Y, Hu J. Diagnostic value of circulating lncRNA ANRIL and its correlation with coronary artery disease parameters. Braz J Med Biol Res 2019;52:e8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yu X, Zou T, Zou L, et al. Plasma long noncoding RNA urothelial carcinoma associated 1 predicts poor prognosis in chronic heart failure patients. Med Sci Monit 2017;23:2226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhang J, Gao C, Meng M, et al. Long noncoding RNA MHRT protects cardiomyocytes against H2O2-induced apoptosis. Biomol Ther 2016;24:19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bank AJ, Rischall A, Gage RM, et al. Comparison of cardiac resynchronization therapy outcomes in patients with New York Heart Association functional class I/II versus III/IV heart failure. J Card Fail 2012;18:373–8. [DOI] [PubMed] [Google Scholar]
- [9].Yang ZJ, Liu J, Ge JP, et al. Prevalence of cardiovascular disease risk factor in the Chinese population: the 2007-2008 China National Diabetes and Metabolic Disorders Study. Eur Heart J 2012;33:213–20. [DOI] [PubMed] [Google Scholar]
- [10].Yuan J, Zou XR, Han SP, et al. Prevalence and risk factors for cardiovascular disease among chronic kidney disease patients: results from the Chinese cohort study of chronic kidney disease (C-STRIDE). BMC Nephrol 2017;18:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen WW, Gao RL, Liu LS, et al. China cardiovascular diseases report 2015: a summary. J Geriatr Cardiol 2017;14:1–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yancy CW, Jessup M, Bozkurt B, et al. 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol 2013;62:e147–239. [DOI] [PubMed] [Google Scholar]
- [13].Tromp J, Ferreira JP, Janwanishstaporn S, et al. Heart failure around the world. Eur J Heart Fail 2019;21:1187–96. [DOI] [PubMed] [Google Scholar]
- [14].Jathar S, Kumar V, Srivastava J, et al. Technological developments in lncRNA biology. Adv Exp Med Biol 2017;1008:283–323. [DOI] [PubMed] [Google Scholar]
- [15].Chen X, Yan GY. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics 2013;29:2617–24. [DOI] [PubMed] [Google Scholar]
- [16].Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform 2016;17:106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochimica et biophysica acta 2014;1839:1097–109. [DOI] [PubMed] [Google Scholar]
- [18].Loewen G, Jayawickramarajah J, Zhuo Y, et al. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol 2014;7:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wu C, Arora P. Long noncoding Mhrt RNA: molecular crowbar unravel insights into heart failure treatment. Circ Cardiovasc Genet 2015;8:213–5. [DOI] [PubMed] [Google Scholar]
- [20].Han P, Li W, Lin CH, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014;514:102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Xuan L, Sun L, Zhang Y, et al. Circulating long non-coding RNAs NRON and MHRT as novel predictive biomarkers of heart failure. J Cell Mol Med 2017;21:1803–14. [DOI] [PMC free article] [PubMed] [Google Scholar]


