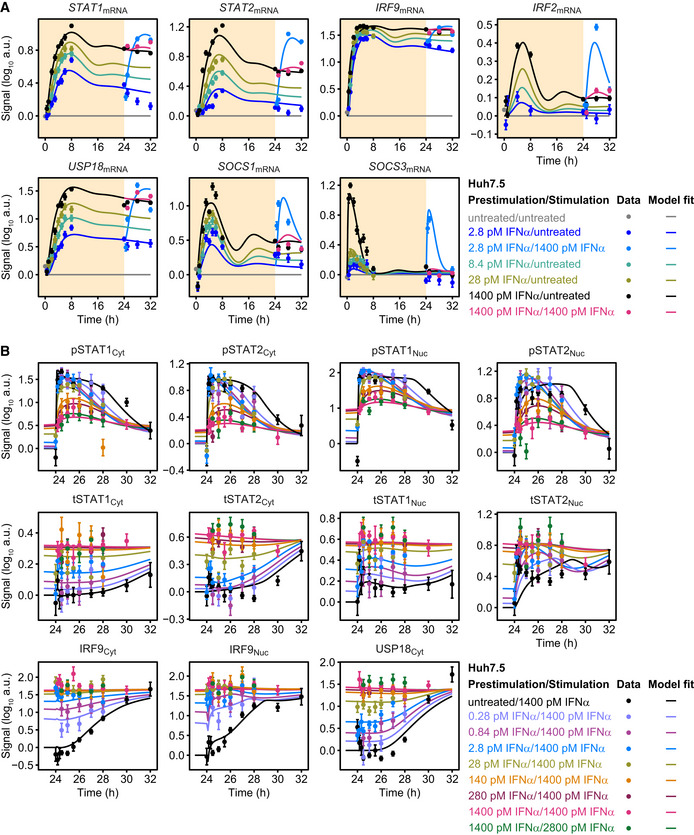
Model calibration with IFNα‐induced expression of feedback transcripts STAT1, STAT2, IRF9, IRF2, USP18, SOCS1, and SOCS3. Growth factor‐depleted Huh7.5 cells were prestimulated with 2.8, 28, and 1,400 pM IFNα (yellow background). After 24 h, cells were stimulated with 1,400 pM IFNα or were left untreated (white background). IFNα‐induced expression of target genes was measured by qRT‐PCR. RNA levels were normalized to the geometric mean of reference genes GAPDH, HPRT, and TBP and were displayed as fold change, visualized by filled circles with errors representing 1σ confidence intervals estimated from biological replicates (N = 3 to N = 14) using a combined scaling and error model. Model trajectories are represented by lines.