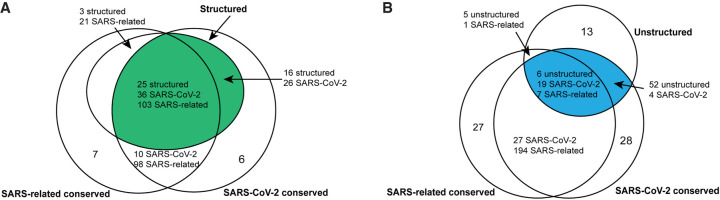
FIGURE 5.
We depict the predicted number of structured, unstructured, and conserved intervals for a choice of sequence conservation cutoffs. The SARS-related conserved intervals are all regions of at least 15 nt with each position at least 90% conserved across an alignment of SARS, bat coronavirus, and SARS-CoV-2 sequences (SARSr-MSA-1). The SARS-CoV-2 intervals are regions of at least 15 nt with each position at least 97% conserved across an alignment of currently available SARS-CoV-2 sequences (SARS-CoV-2-MSA-2). Structured intervals are loci predicted from RNAz with some loci containing multiple RNAz windows, and unstructured intervals are stretches of at least 15 nt where all bases have base-pairing probability at most 0.4. All interval intersections are required to have at least 15 nt overlaps, with the number of overlapping intervals listed for each interval type involved in the intersection. Top-scoring structured intervals conserved in SARS-CoV-2 sequences (green) are listed in Table 2. Top-scoring unstructured intervals conserved in SARS-CoV-2 sequences (blue) are listed in Table 3.