FIGURE 4.
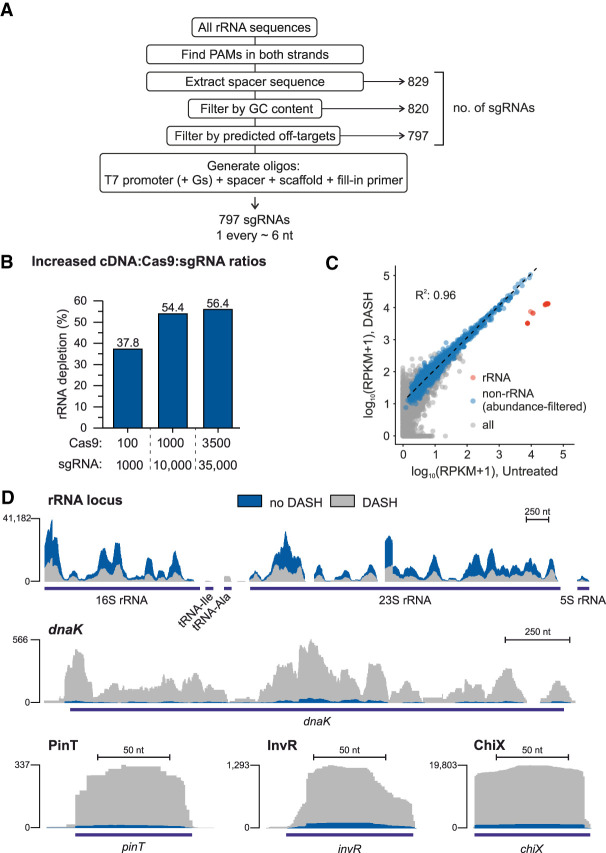
Improved DASH efficiency by maximizing sgRNA target site density. (A) Workflow of the software developed for the design of high-density sgRNA pools. The number of sgRNAs passing each step is indicated to the right. (B) rRNA depletion efficiency using the 797 sgRNA pool in different ratios of Cas9 and sgRNA. Cas9 and sgRNA amounts are indicated as molar excess over a single fragment of the cDNA library. (C) Correlation of transcript abundances in the control versus DASHed (3500:35,000 excess of Cas9:sgRNA) libraries. Red dots (n = 22) represent rRNA transcripts, blue ones (n = 1511) the genetic features with at least 15 reads in the control and 150 reads in the DASH library, and gray dots (n = 3469) the genetic features below this threshold. The regression line and correlation coefficient were computed for the blue dots only. (D) Sequence read coverage of a representative rRNA locus and the dnaK, pinT, invR, and chiX genes in the control and DASH libraries (3500:35,000 excess of Cas9:sgRNA).