FIGURE 6.
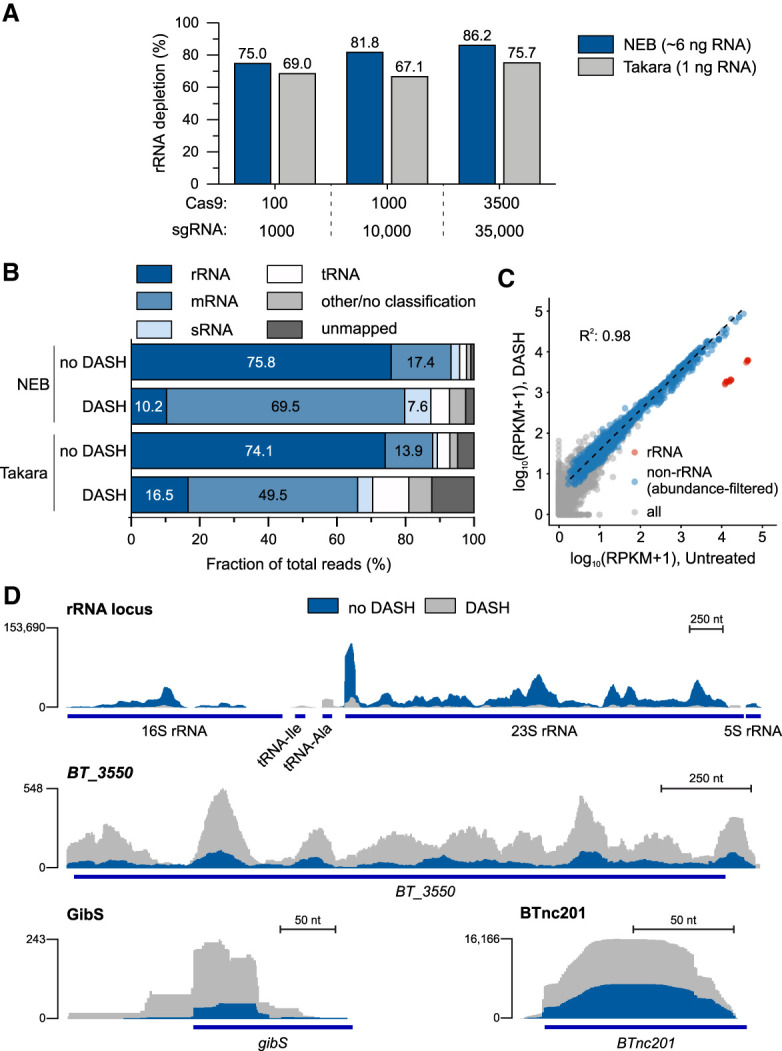
DASH-mediated removal of Bacteroides thetaiotaomicron rRNA. (A) rRNA depletion efficiency from a B. thetaiotaomicron cDNA library. DASH was combined with the different library preparation kits and using the indicated ratios of Cas9 and sgRNA over cDNA fragments. The indicated RNA amounts correspond to the reverse transcription input. (B) RNA class distribution of reads in control and DASH (3500:35,000 excess of Cas9:sgRNA) samples. (C) Correlation of transcript abundances in the control and DASH libraries shown in panel B. Red dots (n = 15) represent rRNA transcripts, blue ones (n = 2474) the genetic features with at least 15 reads in the control library and 150 reads in the DASH sample, and gray dots (n = 2658) the features below this cutoff. The regression line and correlation coefficient were computed for the blue dots only. (D) Sequence read coverage of a representative rRNA locus and BT_3550 (encoding a putative long-chain fatty acid-CoA ligase) and the gibS and BTnc201 regulatory RNA genes (bottom) in the control and DASH library of panels B and C.
