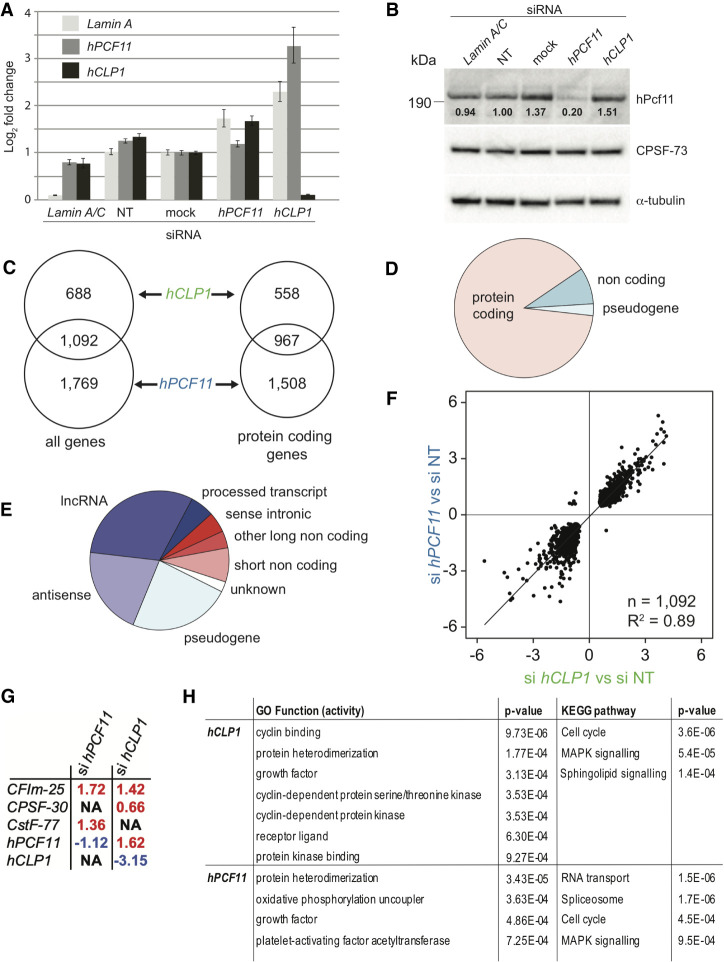
FIGURE 1.
Gene expression analysis of MCF7 cells upon knockdown of hPCF11 and hCLP1. (A) Representative RT-PCR analysis of mRNA levels of Lamin A, hPCF11 and hCLP1 following the indicated siRNA treatments of MCF7 cells. mRNA levels were determined relative to the housekeeping gene ß-tubulin. Lamin A/C siRNA was used as a technical control, nontargeting siRNA (NT) and no-siRNA (mock) served as negative controls. Data are shown as mean ± SD. (B) Western blot analysis of proteins following the indicated siRNA treatments of MCF7 cells. Migration of full-length hPcf11, CPSF-73, and α-tubulin is indicated. Signals for hPcf11 and for α-tubulin were derived from the same PVDF membrane, CPSF-73 was detected on a second membrane. Values below the hPcf11 bands indicate quantification of the intensity of the hPcf11 band relative to α-tubulin. (C) Genomic annotations with significant expression changes following siRNA treatment of MCF7 cells. A total of 1780 significantly differentially expressed annotated features (FDR < 0.05) were identified in hCLP1 knockdown samples (n = 2), and 2861 in hPCF11 knockdown samples (n = 2) compared to nontargeting siRNA control samples (n = 3), 1092 were common to both conditions. A total of 1525 features were annotated to protein coding genes in hCLP1 knockdown samples and 2475 in hPCF11 knockdown samples, with 967 common to both conditions. (D) Breakdown of genomic annotations with expression changes observed upon knockdown of hPCF11 and hCLP1. For genes impacted by both knockdown conditions (1092 genes, FDR < 0.05), expression changes were seen primarily in protein coding genes, 8.5% were noncoding RNA, and 2.7% were pseudogenes. (E) Breakdown of non-protein coding annotations with expression changes observed upon knockdown of hPCF11 and hCLP1. For genes impacted by both knockdown conditions, expression changes were seen in the indicated noncoding RNA species (125 genes, FDR < 0.05). (F) Comparison of expression changes observed upon knockdown of hPCF11 (y-axis) and hCLP1 (x-axis). Scatter plot shows 1092 significantly differentially expressed annotated features (FDR < 0.05) that are found relative to nontargeting siRNA control samples (n = 3) in both hCLP1 knockdown samples (n = 2) and in hPCF11 knockdown samples (n = 2). (G) Expression changes observed with subunits of the core 3′ end machinery upon knockdown of hPCF11 and hCLP1. Significant differential expression levels were determined (FDR < 0.05) and fold changes are indicated in red for increased and blue for decreased expression. NA means that no significant change was observed. (H) Gene ontology of molecular function and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways and associated P-values identified through expression changes observed upon knockdown of hPCF11 and hCLP1. Indicated are all top GO function activity terms and KEGG pathway terms (FDR ≤ 0.05) associated with the knockdown conditions.