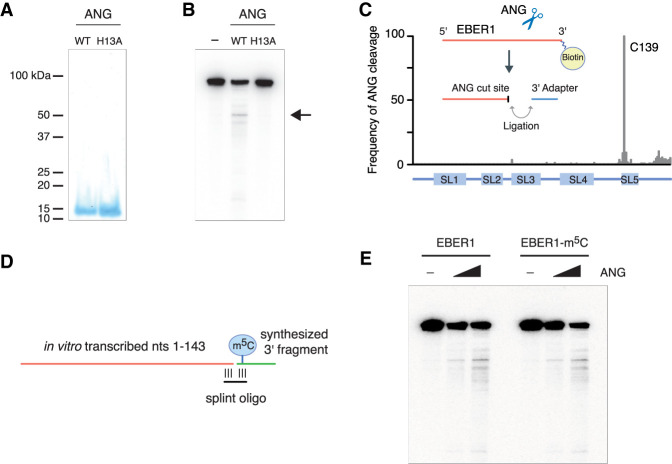
FIGURE 5.
EBER1 is an in vitro substrate of Angiogenin. (A) Recombinant wild-type and catalytically inactive (H13A mutant) recombinant ANG was purified from E. coli. Purified fractions were resolved on a 15% SDS gel and stained with Coomassie. (B) In vitro transcribed EBER1 was incubated with either wild-type or H13A mutant ANG followed by northern blot analysis to detect degradation products of EBER1. While a smear of bands is visible to a minor extent, indicative of random degradation, one preferential cut site within EBER1 is detected (arrow). This degradation product is not observed when incubated with the H13A ANG mutant, demonstrating an ANG-specific RNase activity. (C) EBER1 degradation products were attached to a 3′ linker, converted into an Illumina-compatible library, and deep-sequenced to identify ANG-mediated cut sites. ANG primarily digests EBER1 after C139, which is located in stem–loop 5 adjacent to the m5C site. (D,E) In vitro ANG digestion assay with either unmodified or m5C-modified EBER1 as substrate. Schematic for generating either unmodified or m5C-modified EBER1 by splint ligation. An RNA fragment consisting of EBER1 nucleotides 1–143 was in vitro transcribed using T7 polymerase. A 3′ fragment either lacking or containing m5C145 was chemically synthesized and ligated to the 5′ fragment using a splint oligonucleotide (D). m5C modification of EBER1 does not confer substrate preference in vitro (E).