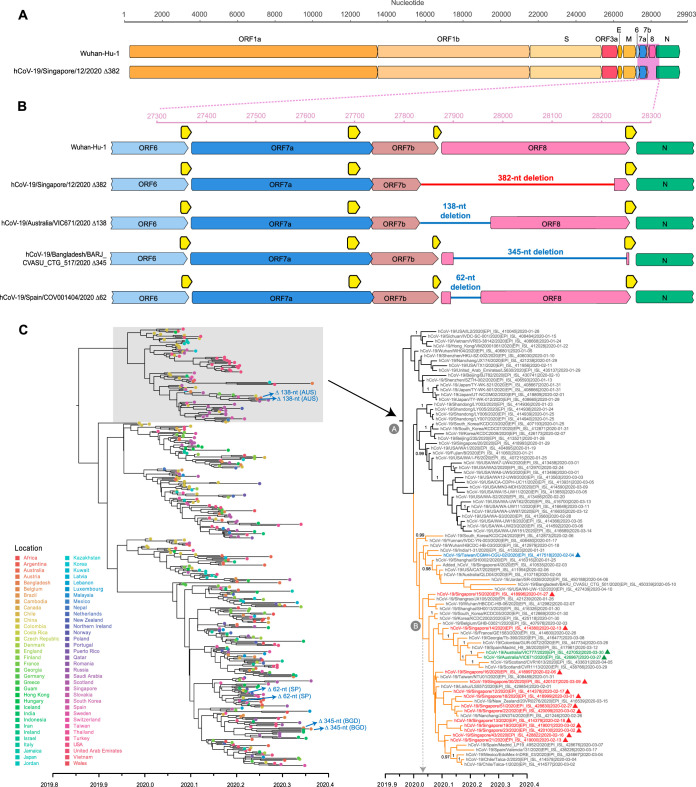
FIG 1.
Genomic organization and evolutionary relationships of human SARS-CoV-2 and SARS-CoV-2 Δ382. (A) Schematic diagram of the genomes of SARS-CoV-2 isolates Wuhan-Hu-1 (GenBank accession no. MN908947) and human CoV-19/Singapore/12/2020 (hCoV-19/Singapore/12/2020) Δ382 (GISAID: EPI_ISL_414378). (B) Magnification of genomic region (pink box in panel A) showing the 382-nt deletion in ORF7b and ORF8 (indicated by red line) in hCoV-19/Singapore/12/2020. Other ORF7b/8 deletions are indicated by blue lines as follows: a 138-nt deletion in hCoV-19/Australia/VIC671/2020 (EPI_ISL_426967), a 345-nt deletion in hCoV-19/Bangladesh/BARJ_CVASU_CTG_517/2020 (EPI_ISL_450344), and a 62-nt deletion in hCoV-19/Spain/COV001404/2020 (EPI_ISL_452497). Horizontal axes indicate the nucleotide position relative to Wuhan-Hu-1; open reading frames (ORFs) are indicated by solid colored arrows. Transcription regulatory sequences (TRSs) are indicated by yellow arrows. (C) Temporal phylogeny of 419 complete genomes inferred using an uncorrelated lognormal relaxed clock model in BEAST. Colored circles at the tips represent geographic locations of virus sampling. Colored triangles represent ORF7b/8 deletion variants. A fully labeled tree with Bayesian posterior probabilities indicated is presented in Fig. S4. Red isolate names indicate SARS-CoV-2 from Singapore with a 382-nt deletion as described in this study. Blue isolate names indicate a 382-nt deletion from Taiwan, whereas green isolate names indicate a 138-nt deletion from Australia. Node A represents the time to most common ancestor (TMRCA) for the lineage containing Δ382 viruses from Singapore and Taiwan, while node B represents the TMRCA of the clade containing Δ382 viruses from Singapore. Bayesian posterior probabilities of ≥0.95 are indicated at nodes. Scale bar represents time in years. Abbreviations: AUS, Australia; SP, Spain; BGD, Bangladesh.