Figure 1: Schematic Representation of the CNV Discrepancy Identification and Resolution Process.
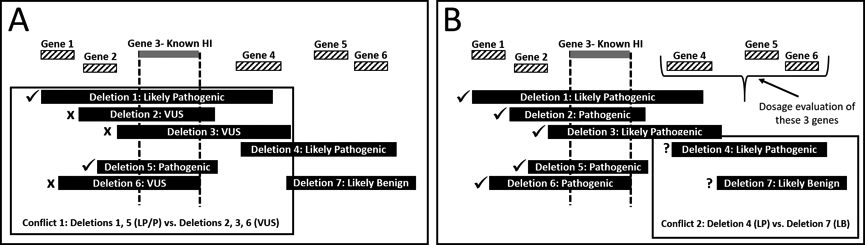
In this generic region of the genome, there are at least two potential conflicts involving CNVs overlapping by ≥50% with different clinical interpretations. A) Conflict 1 involves known haploinsufficient (HI) Gene 3. This information can be used to mediate the conflict resolution process. Likely pathogenic (LP)/pathogenic (P) interpretations are expected for deletions fully encompassing this gene, such as Deletions 1 and 5. Deletions 2, 3, and 6 also fully encompass this gene, but are interpreted as variants of uncertain significance (VUS). These cases would be flagged for reevaluation by the submitting laboratory. B) Assuming Conflict 1 is resolved by the reevaluation process, Conflict 2 can be assessed. To mediate this process, Genes 4, 5, and 6 would be triaged for dosage sensitivity evaluation. The dosage evaluation process could resolve the conflict in and of itself; for example, if Gene 4 were found to be a known HI gene, Deletions 4 and 7 would no longer be in conflict, as Deletion 7 does not include Gene 4.
