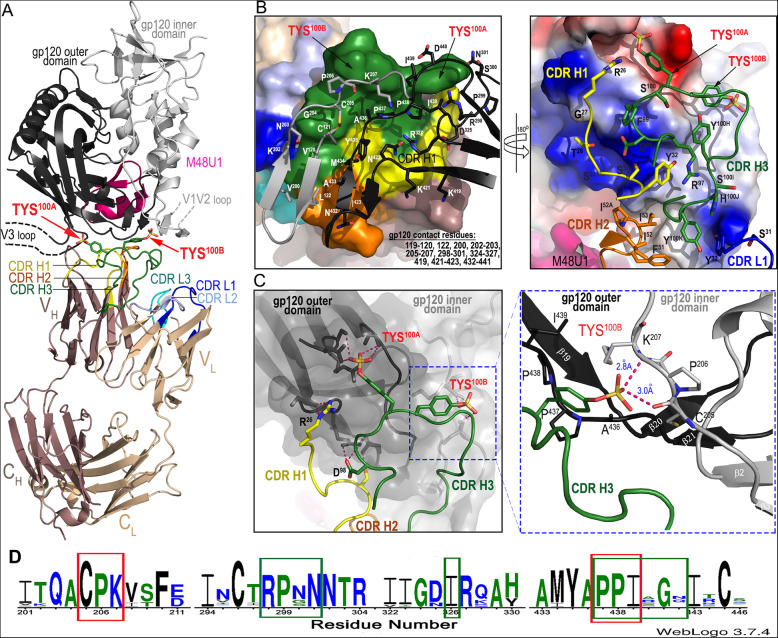
Fig. 1.
Crystal structure of N12-i2 Fab-gp12093TH057 coree-M48U1 complex. a A ribbon diagram of the complex. The gp120 outer and inner domains are black and gray, respectively, with the CDRs of N12-i2 colored as indicated. The two sulfoTyrs essential for N12-i2 binding, TYS100A and TYS100B, are shown as sticks. The V1V2 and V3 loops not present in the gp12093TH057 coree construct are shown as broken lines. b N12-i2 Fab-gp12093TH057 coree interface. The Fab molecular surface colored yellow for CDR H1, orange for CDR H2, green for CDR H3, blue for CDR L1, light blue for CDR L2 and cyan for CDR L3. gp120 contact residues are shown as sticks and outer and inner domains are shown as in a. gp120 contact residues are listed. A 180° view shows details of Fab interaction with the gp120 surface. The gp120 molecular surface is colored based on electrostatic charge, red negative and blue positive. The N12-i2 Fab is displayed as a ribbon diagram with contact residues shown as sticks. Fab contact residues are listed. c Hydrogen bond network formed at the N12-i2 Fab-gp12093TH057 coree interface. The molecular surface is displayed over a gp120 molecule and the gp120 outer and inner domains are shown in black and gray, respectively. The blow-up view shows details of TYS 100B interaction with gp12093TH057 coree. Residues contributing to TYS 100B binding are shown as sticks, and the network of hydrogen bonds is shown with dashed lines. See also Additional file 1: Table S1. d Consensus sequences for gp120 contact residues as defined by a 5-Å cutoff for N12-i2 sulfotryosines 100A (green) and 100B (red) as determined by WebLogo [42]. Approximately 32,000 aligned sequences from the HIV Sequence Database Compendium (https://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/compendium.html) were used as input. Insertions or deletions relative to HXBC2 were excluded for clarity and residue position given relative to HXBC2