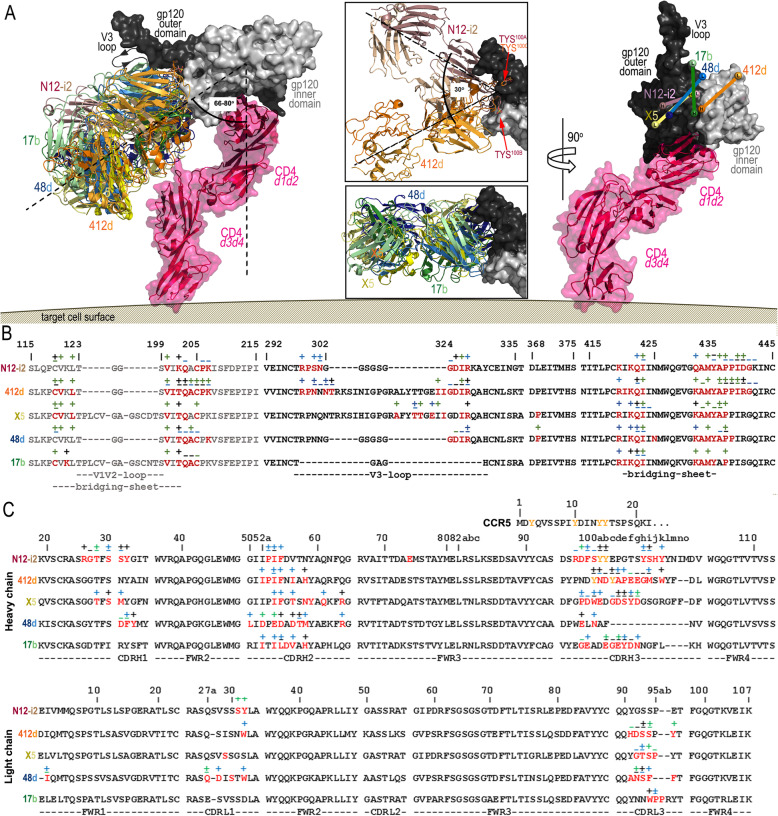
Fig. 4.
Binding of N12-i2 and other CoRBS antibodies to HIV-1 Env. a Overlay of N12-i2 and CoRBS antibodies, 412d (PDB ID 2QAD), X5 (2B4C), 48d (4DVR), and 17b (1GC1), bound to gp120 oriented relative to the target cell membrane. The structural alignment based on the gp120 outer domain of the Fab 412d-gp120YU2 core-CD4 complex (PDB ID 2QAD) and the target cell receptor CD4 extracellular portion modeled by superimposition of the d1-d2 domains to the unliganded CD4 (d1-d4 CD4, PDB code: 1WIO). An alternate conformation of the V3 loop seen in the Fab X5- gp120JR-FL core-CD4 complex (PDB code: 2B4C) is shown as a ribbon. The blow-up views show the alignment from the top (180° vertical rotation showing orientations of N12-i2 and 412d (top) and 48d, X5, and 17b (bottom). Angles were calculated using the center of mass of gp120 and the CD4 d1 domain to define a vector perpendicular to the target cell membrane and the mAb CDR residues to define vectors for the mAbs. A 90° view displays the relative positions of each mAb to CD4-triggered gp120. Heavy- and light-chain (Vh and VL) positions determined by the center of mass of their CDRs (displayed as balls). b Epitope footprints. gp120 buried surface and contact residues shown over the primary gp120 sequence of the isolates used in co-crystallization studies. c Fab interface residues with the N-terminal sequence of the CCR5 co-receptor aligned above the CDR H3 sequence. Tyrosines modified by post-translational O-sulfation are shown in yellow. In b and c, residues buried at the surface as determined by PISA (http://www.ebi.ac.uk/msd-srv/prot_int/cgi-bin/piserver) are shown in red and contact residues as defined by a 5-Å distance cutoff are shown with interactions, main chain (−), side chain (+), or both (±), colored based on contact type: hydrophobic (green), hydrophilic (blue), or both (black)