Figure 2.
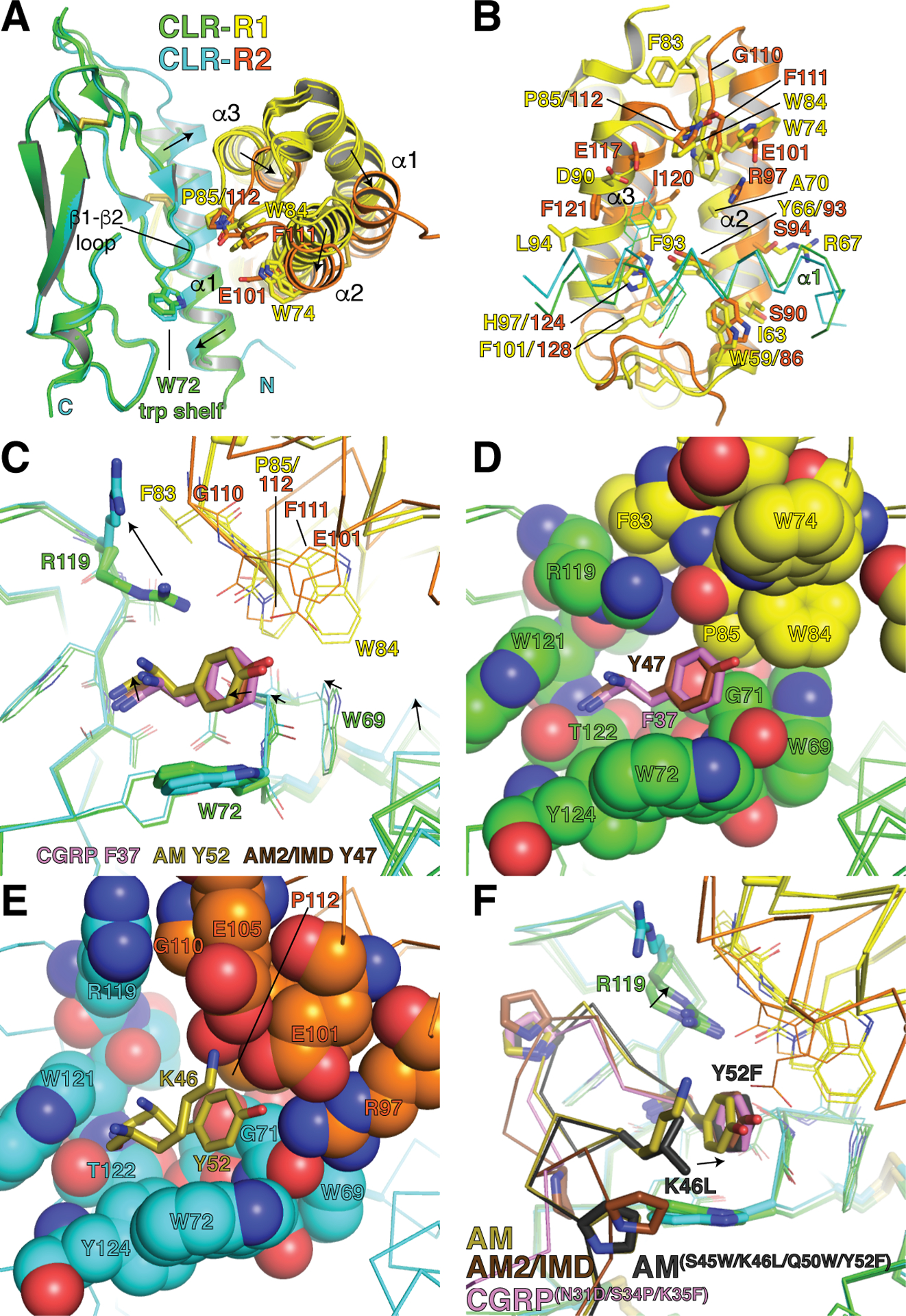
Allosteric modulation of CLR ECD conformation by RAMP1 and RAMP2. (A) Superimpositions of the high-affinity CGRP variant-bound RAMP1-CLR ECD (PDB 4RWG), AM-bound RAMP2-CLR ECD (PDB 4RWF), and AM2/IMD-bound RAMP1-CLR ECD (PDB 6D1U) complexes. For clarity the CLR turret loop is omitted. Arrows indicate shifts occurring from the RAMP1 complexes to the RAMP2 complex. (B) View of the RAMP1/2 α2-α3 interface with the CLR α1 helix (shown as α-carbon trace). (C) Close-up view of the pocket occupied by the peptides’ C-terminal residues, which are shown as sticks. Arrows indicate shifts occurring from the RAMP1 complexes to the RAMP2 complex. (D) View of the pocket in the RAMP1-CLR ECD complexes with residues in space-filling representation. The CGRP F37 and AM2/IMD Y47 C-terminal residues are shown as sticks. (E) View of the pocket in the RAMP2-CLR ECD complex with pocket residues in space-filling representation and the AM K46 and Y52 residues as sticks. (F) Superimpositions of the three structures in panels A and C with RAMP1-CLR ECD bound to a rationally-designed high-affinity, altered selectivity AM variant (PDB 5V6Y). Arrows indicate shifts in the positions of the AM variant C-terminal residue and CLR R119. In all panels the structures were aligned based on the CLR ECD.
