FIGURE 1.
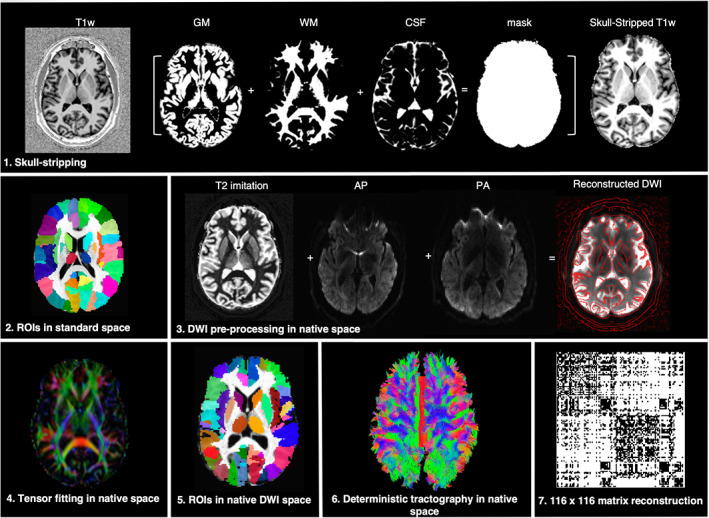
An overview of the imaging processing pipeline. (1) The uniform T1‐weighted image was skull‐stripped by segmenting gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF), multiplying the resulting whole‐brain mask to the image; (2) the anatomical scan was transformed to match the AFNI standard reference template in Talairach‐Tournoux space; (3) motion and eddy current distortions were corrected for both the anterior–posterior (AP) and posterior–anterior (PA) diffusion‐weighted imaging (DWI) data sets using a T2‐weighted imitation image. AP and PA were combined to generate the final DWI reconstruction; (4) the diffusion ellipsoids and parameters were calculated using a nonlinear fitting method; (5) whole‐brain coverage with 116 anatomical regions of interest (ROIs) based on the macrolabel atlas was transferred into individual DWI space; (6) deterministic fiber tracking was performed in the native space between all pairs of ROIs; (7) a 116 × 116 adjacency matrix for each subject was created based on the normalized number of streamlines, thresholded, and used as the measure of structural pairwise connectivity between all ROI pairs (i.e., the nodes of the network)
