Figure 4.
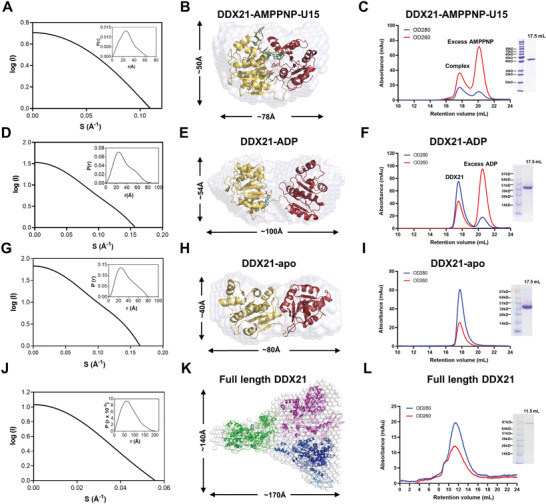
SAXS models and gel‐filtration profiles of full‐length DDX21 and the helicase core. A) The experimental scattering curve and the distance distribution function curve (inset) for DDX21 helicase core (188‐563) with AMPPNP and U15 ssRNA. B) Crystal structure of DDX21‐AMPPNP‐ssRNA (PDB: 6L5N) was fitted into the ab initio envelope obtained from SAXS. C) Gel filtration profile and the SDS‐PAGE of DDX21 helicase core (188‐563) with AMPPNP and U15 ssRNA. The helicase core and U15 ssRNA plus AMPPNP were coeluted out at the retention volume 17.6 mL from the superdex200 10/300 column (GE healthcare, Boston, USA), and excess AMPPNP was eluted out lately. D) The experimental scattering curve and the distance distribution function curve (inset) for DDX21 helicase core (188‐563) with ADP. E) Crystal structure of DDX21 (del‐loop)‐ADP (PDB: 6L5O) was fitted into the ab initio envelope obtained from SAXS. F) Gel filtration profile and the SDS‐PAGE of DDX21 helicase core (188‐563) with ADP. The helicase core with ADP was eluted out at the retention volume 17.5 mL from the superdex200 10/300 column (GE healthcare, Boston, USA), and excess ADP was eluted out lately. G) The experimental scattering curve and the distance distribution function curve (inset) for DDX21 helicase core (188‐563). H) Crystal structure of DDX21 (del‐loop)‐apo (PDB: 6L5L) was fitted into the ab initio envelope obtained from SAXS. I) Gel filtration profile and the SDS‐PAGE of DDX21 helicase core (188‐563). The helicase core was eluted out at the retention volume 17.6 mL from the superdex200 10/300 column (GE healthcare, Boston, USA), corresponding with a molecular weight around 30 kD, indicating the possible monomer status in solution. J) The experimental scattering curve and the distance distribution function curve (inset) for full‐length DDX21. (K) The modeled structure of full‐length DDX21 was fitted into the SAXS ab initio envelope with three DDX21 molecules. The modeled DDX21 structure was obtained using the Phyre2 web portal (http://www.sbg.bio.ic.ac.uk/~phyre/). L) Gel filtration profile and the SDS‐PAGE of full‐length DDX21 (1‐783). DDX21 was eluted out at the retention volume 11.6 mL from the superdex200 10/300 column (GE healthcare, Boston, USA), corresponding with a molecular weight around 340 kD, indicating the possible trimer or tetramer status in solution.
