Figure 5.
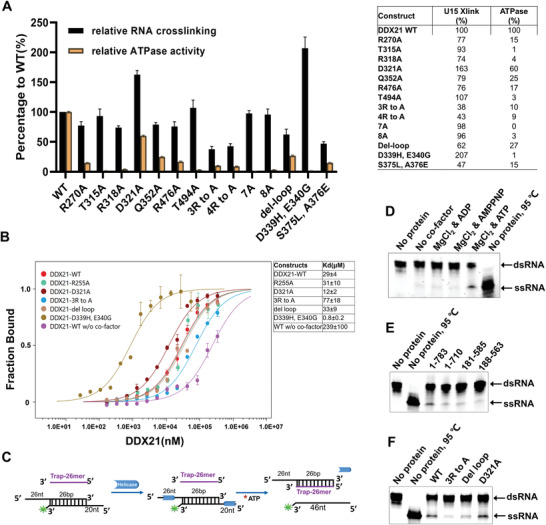
Mutational analysis of key residues involved in ATP hydrolysis and RNA binding. A) Histogram (left panel) and detailed values (right panel) of the RNA‐crosslinking and RNA‐stimulated ATPase activities, which are shown as percentages of the wide‐type DDX21 activity. All presented data represent means ±S.D. of three independent determinations. B) The U15 binding affinities of WT and mutant DDX21. Error bars represent the standard error between three replicate experiments. C) Schematic diagram of the unwinding process with 5’‐tailed RNA duplex and trap RNA. D) Full‐length DDX21 unwinds RNA duplex with or without different cofactors. The final products of unwinding reaction were separated by a 4–20% gradient Novex TBE Gel (Thermo Fisher Scientific, Waltham, USA). DDX21 unwound 5′‐tailed RNA duplex effectively with ATP and MgCl2, while very weak bands of ssRNA were still observed from other reaction pools without ATP. E) RNA unwinding with full length and different truncated forms of DDX21 by using a 4–20% gradient Novex TBE Gel (Thermo Fisher Scientific, Waltham, USA). The unwinding reactions were proceeded with 2 × 10−3 m ATP and 2 × 10−3 m MgCl2 by using 2 × 10−6 m DDX21 full‐length or truncations, 0.2 × 10−6 m 5′‐tailed RNA duplex, and 0.8 × 10−6 m unlabeled trap RNA. F) RNA unwinding with the full‐length WT and mutant DDX21, by using a 4–20% gradient Novex TBE Gel (Thermo Fisher Scientific, Waltham, USA), and the same reaction conditions described above in (E).
