Figure 7.
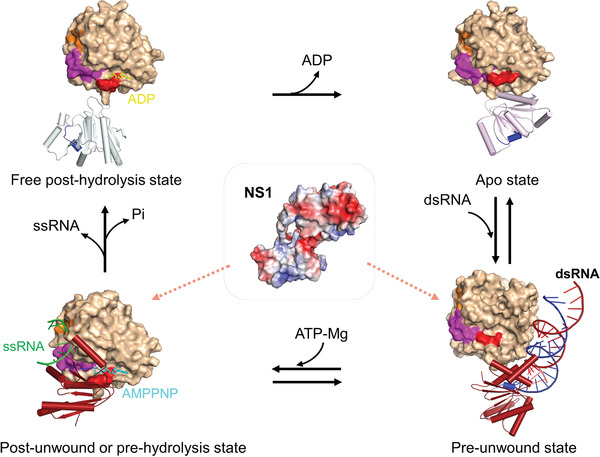
A proposed model for DDX21 unwinding cycle associated with NS1. A four‐step unwinding cycle represented by the apo‐ (PDB: 6L5L, this work), pre‐unwound (PDB: 6O5F), post‐unwound (PDB: 6L5N, this work), and post‐hydrolysis (PDB: 6L5O, this work) states. Full‐length NS1 might bind to DDX21 through RNA in its pre‐unwound state or post‐unwound state. The D1 of all the structures are illustrated as a molecular surface (light orange), and the D2 domains are shown as cartoon models (helices as cylinders, strands as arrows, and loops as tubes). On the D1 surface, the orange color represents motif Ic (the “wedge” helix); the magenta color represents the “DF loop” spanning from DEVD box within motif II to the residue Phe349; the red color represents the “TG loop” within the motif I; the blue color in D2 domain represents the motif V (the “sensor”). DDX3X‐dsRNA structure (PDB: 6O5F) is used here to represent DDX21 pre‐unwound state and only one DDX3X molecule is shown for clarity. The corresponding motifs in the DDX3X structure are shown in the same colors as for DDX21. All these motifs within both D1 and D2 have significant conformational changes during the unwinding cycle.
