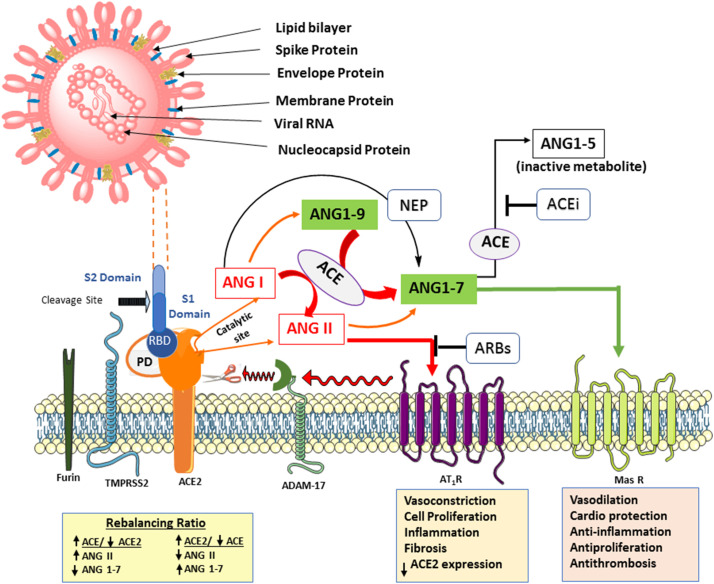
Fig. 1.
SARS-COV-2 virus binding ACE2 and the renin-angiotensin system axis.
Top left shows the virus structure identifying protruding surface spike proteins (enlarged to show S1 and S2 segments with the Receptor Binding Domain (RBD)), the most abundant Membrane protein, the Envelope protein that forms a simple multimeric ion channel, and the Nucleocapsid protein with the bound positive sense single strand (ss) RNA. Below: Furin pre-cleaves spike proteins at the S1/S2 site and promotes subsequent TMPRSS2-dependent entry into host cells. The RBD in the S1 segment binds with high affinity the protease domain (PD) in ACE2. Subsequent conformational changes in S2 facilitate fusion between the viral envelope and the host cell membrane, and internalization by endocytosis with ACE2. Ang II-AT-1 receptor activation of ADAM17 cleaves/sheds cell surface ACE2 which can no longer cleave Ang II or Ang I to protective peptides (Ang (1-9) and Ang (1-7)), resulting in myocardial dysfunction. This action is blocked by AT-1 receptor blockers upregulating ACE2 levels. The AT-1 receptor-mediated effects are opposite to that of Mas and MrgD (not shown) receptors. The catalytic subunit of ACE2 is independent of PD and may operate as long as ACE2 is not internalized. The protective axis comprising of recombinant ACE2, Ang (1–7), Mas receptor agonists, and RAS inhibitors enhance ACE2 action and serve as potential therapies in Covid-19. In addition, ACE inhibitors prevent the metabolism of Ang (1-7) by ACE to the inactive metabolite Ang (1-5) (see Table 1).