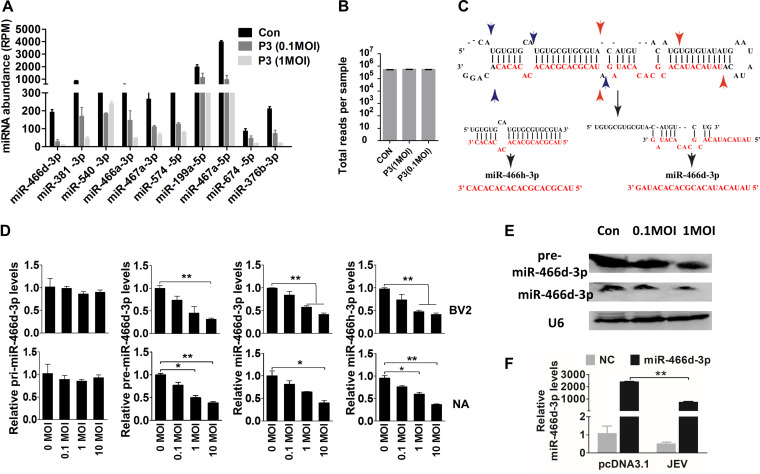
FIG 2.
JEV infection induces incorrect processing of host miRNAs. (A and B) Sequencing of multiplexed miRNA after JEV infection. (A) Read numbers of miRNAs in NA cells infected with JEV at an MOI of 0.1 or 1 or mock infected. (B) Total numbers of reads per sample. (C) Schematic of pre-miR-466d-3p processing and types of mature miRNAs (miR-466d-3p and miR-466h-3p). The black arrows indicate the processing direction of miRNA, the blue arrowheads represent miR-466h-3p located in pre-miR-466d, and the red arrowheads represent miR-466d-3p located in pre-miR-466d. (D) Expression of mature miR-466d-3p, miR-466h-3p, pre-miR-466d-3p, or pri-miR-466d-3p using qRT-PCR in NA or BV2 cells infected with P3 at the indicated MOI after 48 h of infection. (E) Detection of miRNA by Northern blotting. Total RNA was isolated with RNAiso Plus and detected with a DIG-labeled probe. (F) Quantification of degradation of exogenous miR-466d-3p mimic in NA cells by qRT-PCR. NA cells were infected with P3 at an MOI of 0.1 and transfected with miR-466d-3p at 36 hpi. After 48 h of infection, the total RNA from NA cells was quantified. For all graphs, results are shown as means and SD. Significance was assessed using Student's t test. *, P ≤ 0.05; **, P ≤ 0.01.