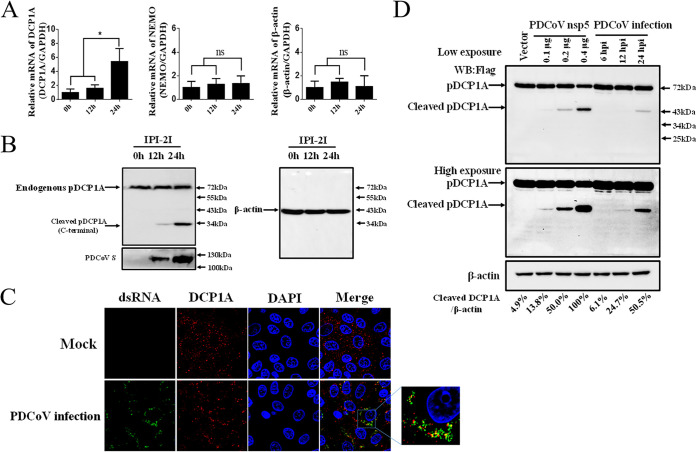
FIG 3.
PDCoV infection cleaves endogenous pDCP1A. (A) PDCoV strain CHN-HN-2014 was used to infect IPI-2I cells (MOI = 0.5) and then harvested to detect pDCP1A mRNA by RT-qPCR at 0, 12, or 24 h postinfection (hpi). (B) IPI-2I cells were infected with PDCoV as described for panel A. Endogenous pDCP1A was detected by Western blotting with antibody against human DCP1A (hDCP1A) at 0, 12, and 24 hpi. (C) IPI-2I cells were infected with PDCoV (MOI = 0.5). At 24 hpi, cells were fixed and then stained with mouse monoclonal antibody specific for dsRNA and rabbit monoclonal antibody against DCP1A, followed by incubation with Alexa Fluor 488-conjugated donkey anti-mouse IgG antibody (green) and 594-conjugated donkey anti-rabbit IgG antibody (red). Nuclei were stained with DAPI (blue). (D) IPI-2I cells were cotransfected with pCAGGS-Flag-pDCP1A and various doses of PDCoV nsp5 expression plasmid. In parallel, IPI-2I cells were transfected with 2.5 μg of pCAGGS-Flag-pDCP1A and then infected with PDCoV (MOI = 0.5). The cotransfected cells were collected at 28 h posttransfection, and the infected cells were collected at different time points (6, 12, 24 hpi) for Western blotting. Quantification of the Western blots was performed by grayscale value analysis with ImageJ software, and the relative cleaved DCP1A data were normalized to those for β-actin. *, P < 0.01; ns, not signficant.