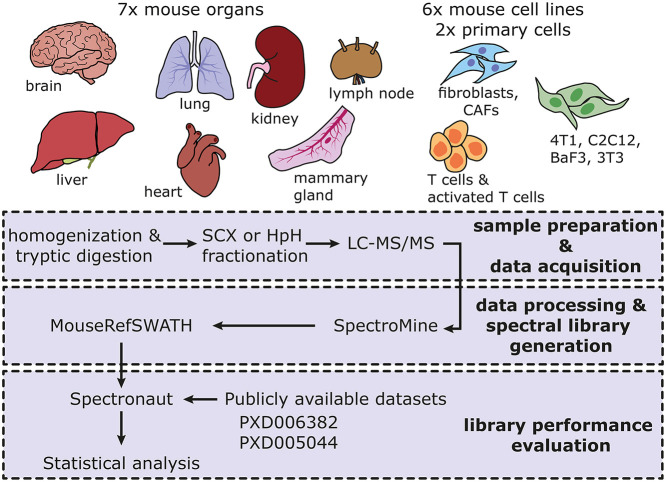
Fig. 1.
Overview of the sample types and schematic workflow used to build the MouseRefSWATH reference spectral library. Fifteen unique sample types comprising seven mouse organs, two primary cell types and six mouse cell lines were analysed to maximize proteome coverage. The workflow consists of a first step of sample preparation and data acquisition of each sample by data-dependent acquisition (DDA)-based liquid chromatography–tandem mass spectrometry (LC-MS/MS). In the second step, the resulting proteomic data were subjected to processing and spectral library generation using the combined Search Archives approach in the SpectroMine software. The subsequent evaluation of the performance of the MouseRefSWATH library was undertaken with the Spectronaut software utilizing two publicly available datasets. CAF, cancer-associated fibroblast; HpH, high pH; SCX, strong cation exchange.