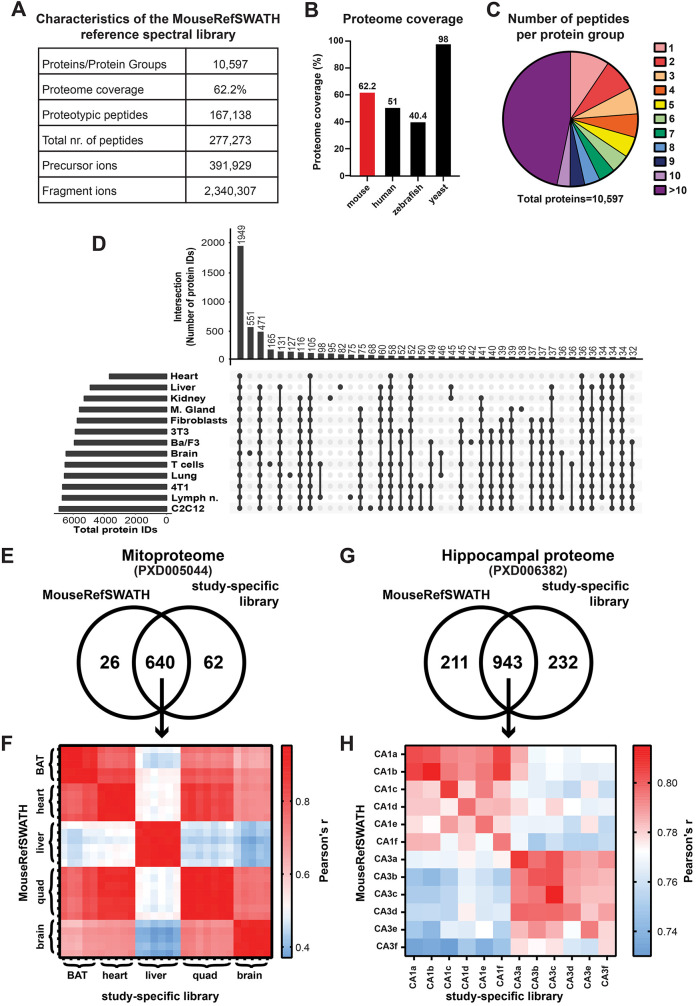
Fig. 2.
Characteristics and performance of the MouseRefSWATH reference spectral library. (A) Detailed characteristics of the generated MouseRefSWATH library. (B) Percentage proteome coverage of MouseRefSWATH library in comparison with published spectral libraries for other eukaryotic organisms, including human (Rosenberger et al., 2014), zebrafish (Blattman et al., 2019) and yeast (Picotti et al., 2013). (C) Peptide coverage of the individual proteins in the MouseRefSWATH library. 90.6% of the proteins are represented by more than one unique peptide. (D) Contribution plot of the tissue- or cell-specific proteins to the overall composition of the MouseRefSWATH library. The bar chart at the top depicts the total number of proteins identified in the overlap of datasets contributed by different organs/cell types as indicated in the dot plot below. The bar chart on the left indicates the total number of proteins identified in the individual datasets from each organ or cell type. (E) Venn diagram depicting overlap of mitochondrial proteins detected by the MouseRefSWATH library and study-specific library based on analysis of the PXD005044 dataset (Williams et al., 2018). (F) Similarity matrix showing Pearson's correlation coefficient of the 640 overlapping mitochondrial proteins, which were quantified by either the MouseRefSWATH library or the study-specific library. Five to eight animals were used for each tissue site. BAT, brown adipocyte tissue; quad, quadriceps. (G) Venn diagram depicting overlap of hippocampal proteins detected by the MouseRefSWATH library and study-specific library based on analysis of the PXD006382 dataset (von Ziegler et al., 2018). (H) Similarity matrix showing Pearson's correlation coefficient of the 943 overlapping hippocampal proteins, which were quantified by either the MouseRefSWATH library or the study-specific library. Two hippocampal areas (CA1 and CA3) from six animals (a-f) were used.