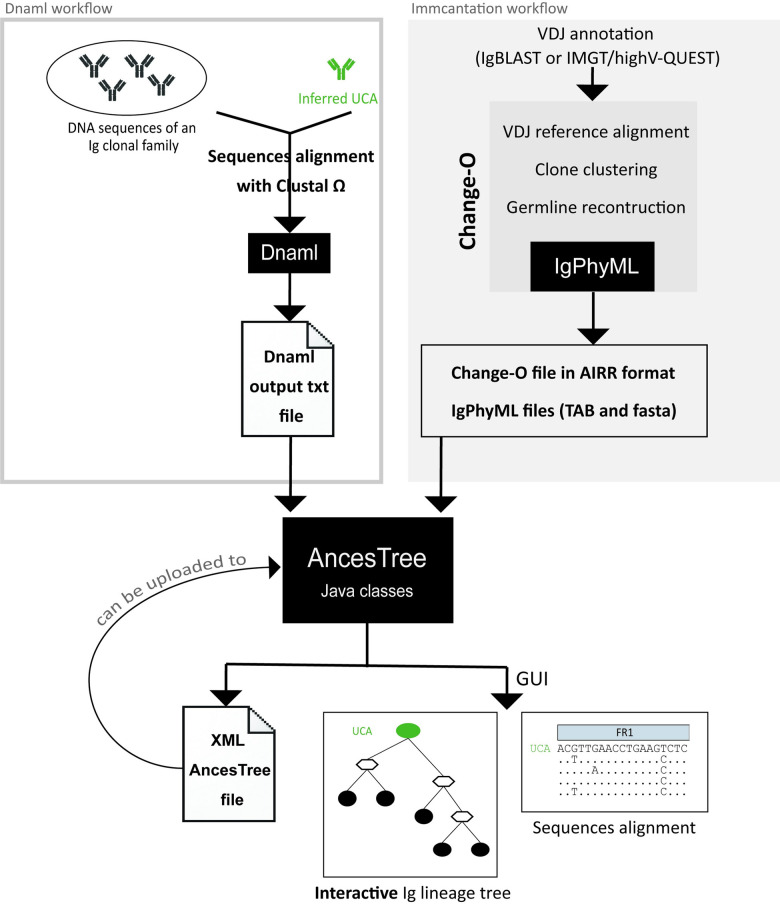
Fig 1. AncesTree workflow.
Dnaml workflow is used (left part of the figure), DNA sequences are aligned with Clustal Ω and processed by Dnaml, a phylogenetic tree is generated. If the Immcantation workflow is used (right part of the figure), reads are processed through Change-O pipeline and IgPhyML will generate the trees. AncesTree processes the different inputs and reconstructs the phylogenic tree with all information related to Ig. The tree is displayed in a GUI and an Extensible Markup Language (XML) file is produced (that could be used as direct input into AncesTree).