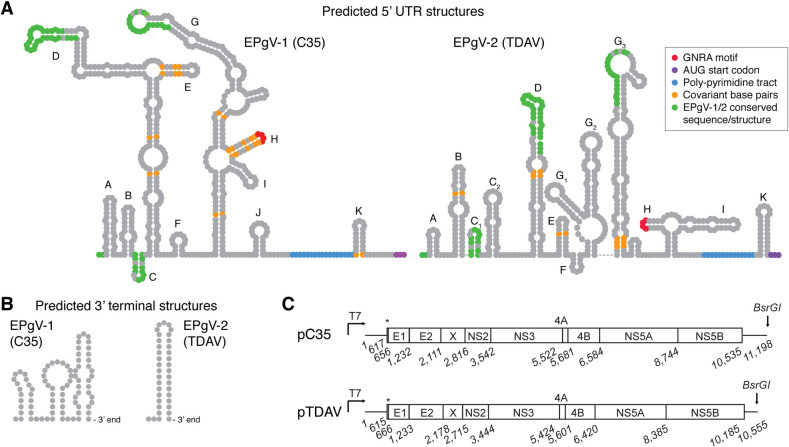
Fig 3. Structures of EPgV RNA ends and consensus clone genomes.
(A) Prediction of RNA secondary structures of the C35 and TDAV 5’ UTRs were performed using MFOLD [59] and guided by covariant base-pairs from alignment of isolates. Stem-loops are labelled according to previous EPgV-1 structure prediction [12]; corresponding labelling of TDAV stem-loops is attempted. Only little direct similarity is evident between 5’ UTR structures of EPgV-1 and -2; this is indicated in green. Other features are colored as indicated. (B) Predicted 3’ UTR terminal stem-loops. (C) Schematic outline of the pC35 and pTDAV molecular consensus clones drawn to scale with nucleotide numbering of junctions predicted from alignment to other pegiviruses and using SignalP 5.0 [60]. Asterisks indicate the N-terminal peptide of the polyprotein. High resolution structures and alignments of UTRs are shown in S2 and S3 Figs.