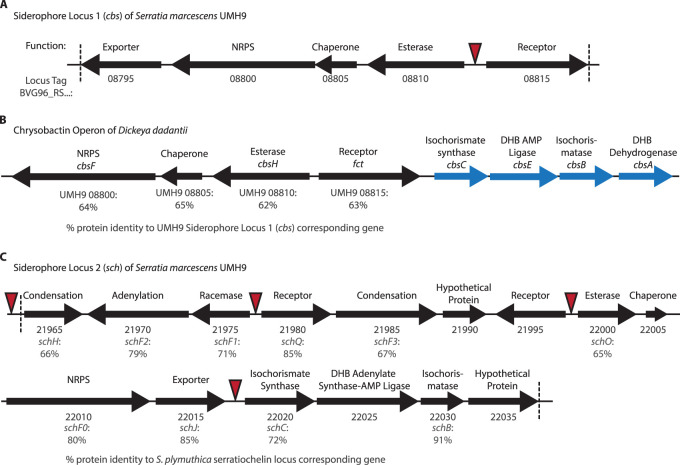
FIG 2.
Siderophore locus 1 (cbs) is similar to the chrysobactin operon in Dickeya dadantii, while siderophore locus 2 (sch) shows similarity to Serratia plymuthica serratiochelin genes. (A) The UMH9 cbs locus has an organization similar to that of the chrysobactin operon found in Dickeya dadantii (B). Four genes (in blue) are missing from the UMH9 cbs locus that are present in the D. dadantii chrysobactin operon. However, these genes, involved in the production of chrysobactin precursor molecules, are found elsewhere in the UMH9 genome. The percentages reported underneath the genes in panel B are the protein percent identity to the UMH9 siderophore locus 1 corresponding gene in panel A. (C) The UMH9 sch locus contains genes predicted to be necessary for siderophore biosynthesis and transport. The percentages reported under the genes are the amino acid percent identity to the S. plymuthica serratiochelin locus corresponding gene. The gene function is noted above each arrow. The UMH9 locus tag number (BVG96_RS…) is referenced below each arrow for panels A and C. Red triangles indicate the locations of putative ferric uptake regulator (Fur) binding sequences in the UMH9 genome. The genes located between the vertical hashed lines in panels A and C were deleted in the cbs::kan and sch::kan mutants, respectively, as well as in the cbs::cam sch::kan mutant together.