Figure 3. γ-TuRC localizes to RAB-11.1 endosomes.
(A) Electron micrograph of the dendritic growth cone. Red arrows, MTs; white arrowhead, clear-core vesicle; scale bar, 0.2 μm. (B) Reconstruction of serial electron microscopy sections through the anterior dendrite. Yellow lines, MTs in the distal dendrite; green lines, MTs crossing the predicted MTOC region; red lines, MTs in the proximal region; white circles, clear-core vesicles; blue circles, dense-core vesicles; scale bar, 1 μm. (C) MT number distribution determined by serial electron microscopy in the anterior dendrite growth cone region in three PVD neurons during development. (D) GIP-2 and RAB-11.1 colocalization in the dendritic growth cone region. Scale bar, 5 μm. (E) Quantification of GIP-2 and RAB-11.1 fluorescence overlap in the growth cone region (n = 11 individual animals). ****p<0.0001, Brown-Forsythe and Welch ANOVA test, error bars represent SEM. (F) Normalized intensity of GIP-2 and RAB-11.1 from the dendritic tip along the dendrite shaft to the cell body at different time points. Black arrow, direction of GIP-2 and RAB-11.1 movement. Vertical dashed line, the center of GIP-2 and RAB-11 endosomes at t=0s. (G) Kymograph of GIP-2::GFP and mCherry::RAB-11.1 in the growth cone region. Horizontal scale bar, 2 μm; vertical scale bar, 10 s. (H) GIP-2 localization in worms overexpressing RAB-11.1(S25N) dominant negative mutant. (I) GIP-2 localization in rab-11PVD(-) worms (Punc-86::Cre; rab-11.1(wy1444[lox])): multiple dim GIP-2 puncta in cell body (i1 and i2), dispersed dim GIP-2 puncta along the dendrite shaft (i3). Dashed white lines: PVD outline; white arrows, GIP-2 puncta; gray arrows, unrelated signal from gut granules. (J) Quantification of GIP-2::GFP class of localization in wt worms, worms overexpressing a RAB-11, RAB-11(S25N) dominant negative mutant or rab-11PVD(-) worms (Punc-86::Cre; rab-11.1(wy1444[lox])). Scale bar, 5 μm. A, anterior; P, posterior. Images in D and H were taken in the glo-1(zu391) mutant background to reduce the gut granule signal.
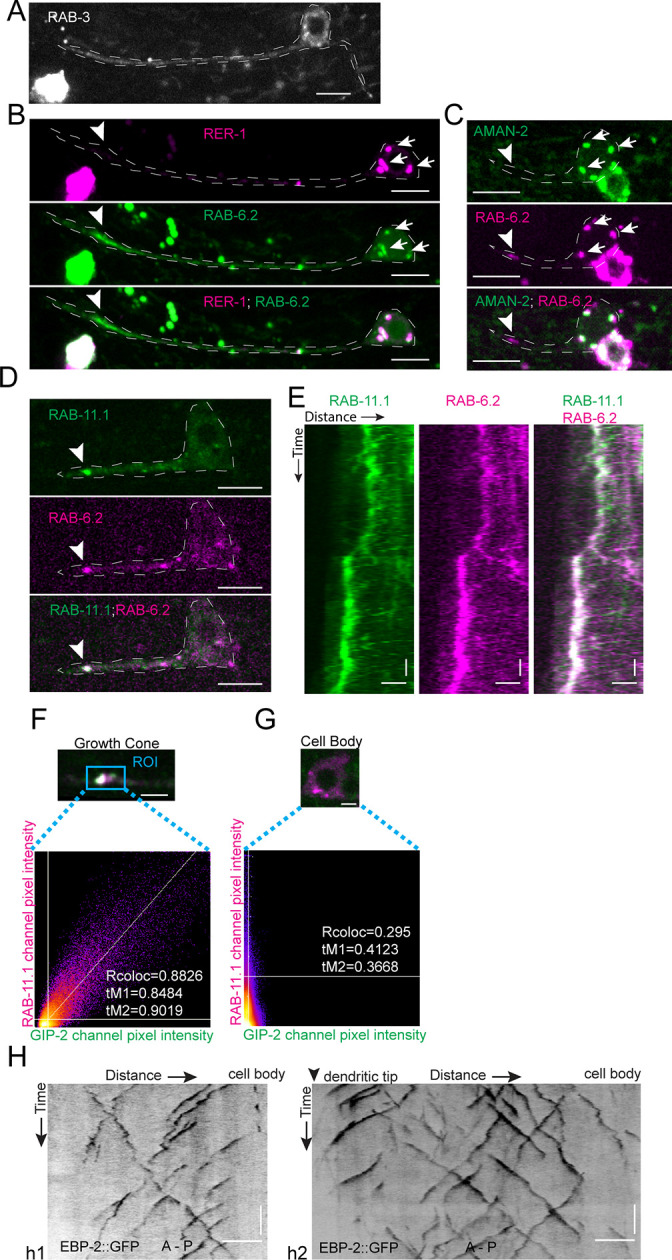
Figure 3—figure supplement 1. Synaptic vesicles and Golgi stacks and outposts do not localize to the growth cone region.