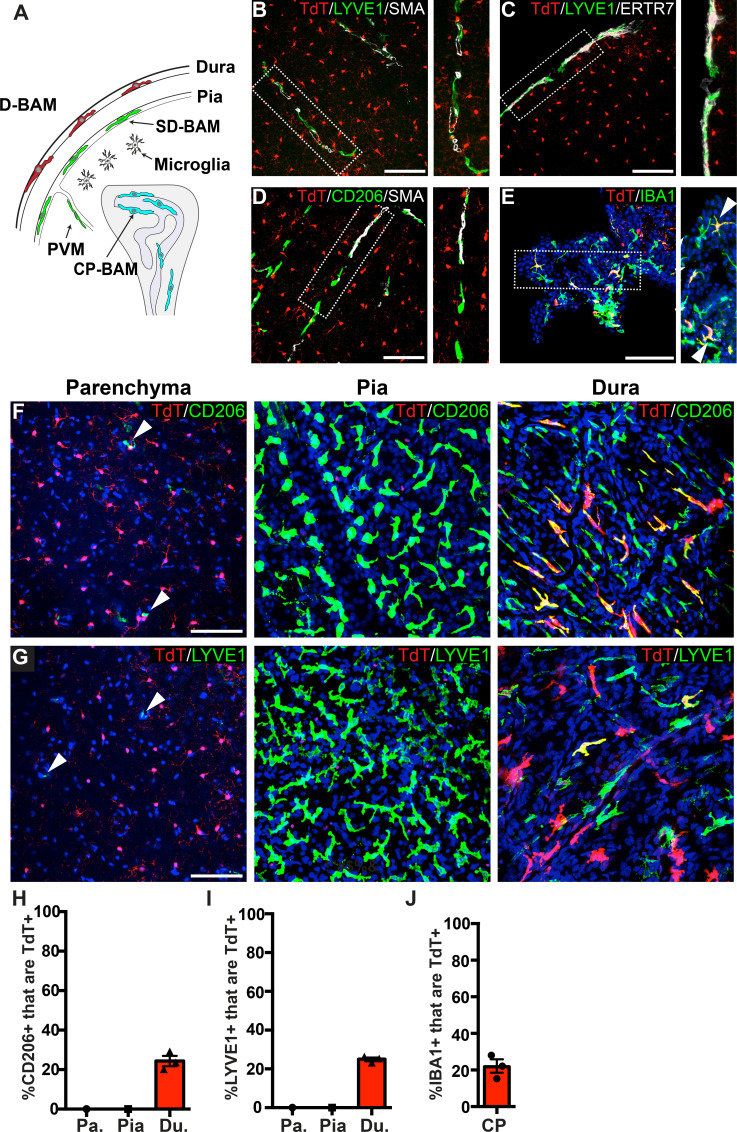
Figure 2. Analysis of P2ry12-CreER recombination in brain macrophage populations.
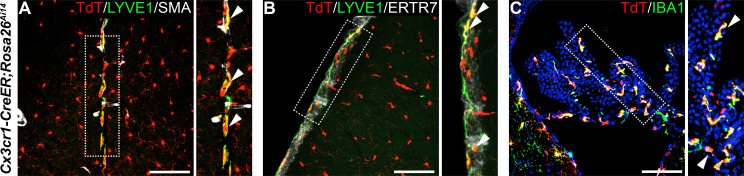
(A) Cartoon depicting the subsets of brain macrophage populations that were analyzed in P2ry12-CreER; Rosa26Ai14 mice. D-BAMs, SD-BAMs, CP-BAMs = dural, subdural, choroid plexus border-associated macrophages. PVMs = perivascular macrophages. Adapted, with permission, from Van Hove et al., 2019. (B) Analysis of P2ry12-CreER; Rosa26Ai14 recombination in SMA-adjacent LYVE1+ perivascular macrophages. No recombination was seen in perivascular macrophages, unlike Cx3CR1-CreER; Rosa26Ai14 mice (see Figure 2—figure supplement 1). (C) Analysis of recombination in LYVE1+ pial macrophages. ERTR7 expression delineates pial boundaries. (D) Analysis of recombination in SMA-adjacent CD206+ perivascular macrophages. (E) Analysis of recombination in IBA1+ macrophages of the choroid plexus. (F) Whole-mount cerebral cortex and dura, immunostained for CD206. (G) Whole-mount cerebral cortex and dura, immunostained for LYVE1. Weakly red cells in pial images are microglia in deeper focal planes. (H–I) Quantification of recombination in CD206+ (H) and LYVE1+ (I) macrophages in the parenchymal, pial and dural spaces. (J) Quantification of recombination in IBA1+ macrophages of the choroid plexus. Error bars = SEM. Scale bars = 100 µm (B–G).