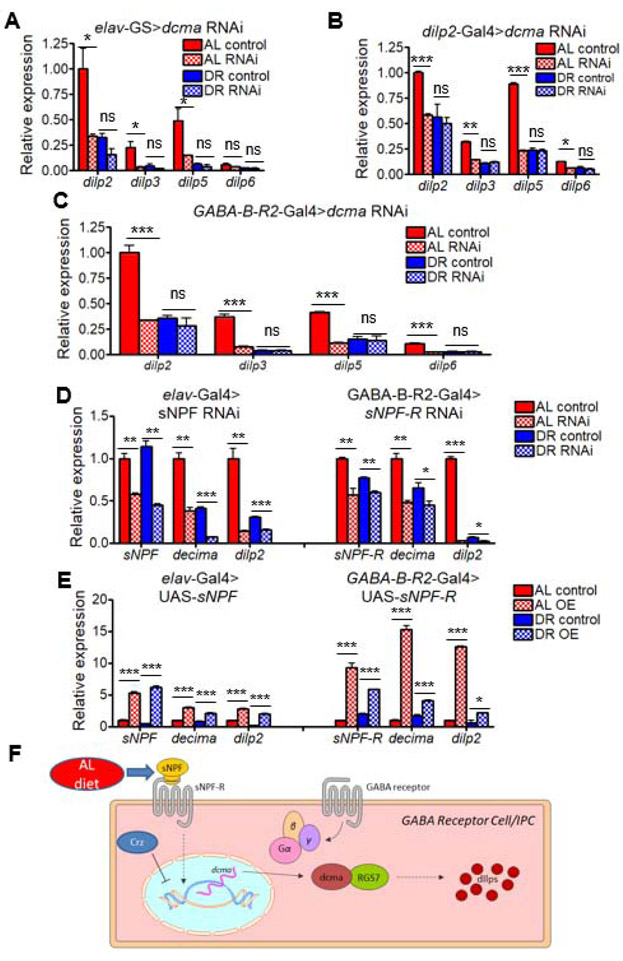
Figure 6. decima regulates insulin-like peptide production in GABA receptor neurons through sNPF signaling.
(A-C) Levels of neuronally-expressed dilps in heads of flies with dcmaRNAi driven (A) pan-neuronally under control of RU486, (B) in dilp2-producing cells, and (C) in GABA receptor neurons. (D) Relative expression of dcma and dilp2 with pan-neuronal RNAi of sNPF (left) or GABA Receptor cell-specific sNPF-RRNAi (right) in heads of flies relative to rp49 housekeeping gene. (E) Relative expression of dcma and dilp2 with pan-neuronal overexpression of sNPF (left) or GABA Receptor cell-specific sNPF-R overexpression (right) in heads of flies relative to rp49 housekeeping gene. AL shown in red, DR in blue. RNAi represented by checkered bars and control conditions in solid bars. N = 50 fly heads per condition. Significant differences between RNAi and controls are indicated by *. * = p < 0.05, ** = p < 0.005, *** = p < 0.0005, determined by unpaired t test. p values shown in Data S3. nc = no change, ns = not significant. N = 200 flies per condition for each RNAi experiment. All experiments show collective results of three biological replicates. Error bars represent SD between replicates. (F) Model for decima’s regulation of dilp production. dcma transcription is upregulated through dietary and sNPF signals, and in turn then regulates the transcription of Drosophila insulin-like peptides in the GABA receptor neurons/insulin-producing cells. See also Figure S7.